With its intricate network of specialized blood arteries forming a nearly impenetrable barrier, the brain is the most protected organ in the body. This special anatomical configuration shields it from external attackers, but it also makes it challenging for researchers to investigate the expression of individual genes and how variations in gene expression can result in disease.
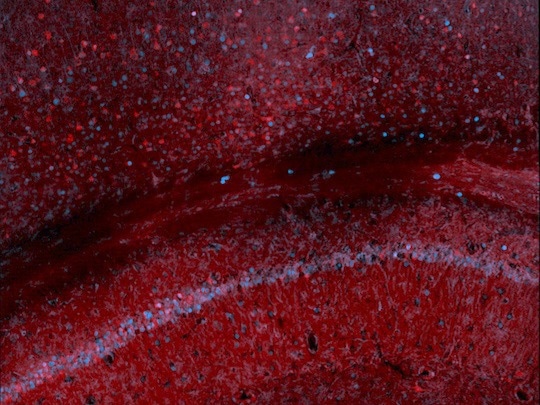 Scan of mouse hippocampus and cortex, where the red signal derives from immunostaining for the synthetic marker developed by the Szablowski lab and the blue from the immunostaining of c-Fos, an endogenous protein whose production is heightened by increased neuronal activity. Image Credit: Szablowski lab/Rice University.
Scan of mouse hippocampus and cortex, where the red signal derives from immunostaining for the synthetic marker developed by the Szablowski lab and the blue from the immunostaining of c-Fos, an endogenous protein whose production is heightened by increased neuronal activity. Image Credit: Szablowski lab/Rice University.
According to a paper published in Nature Biotechnology, researchers at Rice University have created a non-invasive method of tracking the dynamics of gene expression in the brain, which will facilitate future research into neurological disorders, cognitive function, and brain development.
Released markers of activity (RMAs) are a novel class of molecules that Rice bioengineer Jerzy Szablowski and colleagues have created. These molecules can be used as a simple blood test to detect gene expression in the brain.
Typically, if you wanted to look at gene expression in the brain, you would have to wait to do a post-mortem analysis. There are some more modern neuroimaging techniques that can do this, but they lack sensitivity and specificity to track changes in specific cell types. With the RMA platform, we can introduce a synthetic gene expression reporter to the brain, which produces a protein that can pass through the blood-brain barrier. We can then measure changes in expression for a gene of interest with a simple blood test.”
Jerzy Szablowski, Assistant Professor, Department of Bioengineering, George R. Brown School of Engineering, Rice University
Jerzy Szablowski saw that the brain would rapidly clear injections of antibody treatment. Thus, he initially considered the prospect of a synthetic gene expression reporter.
Whenever these injections were done, the antibodies would just disappear ⎯ they wouldn’t hang around long enough in the brain for effective therapy, but we thought the failure of antibody therapies could be repurposed to our advantage. What if we took the part of the antibody responsible for this escape and attached it to a protein that could be easily detected? We could then see where, when, and how much of a particular gene was being expressed in the brain.”
Jerzy Szablowski, Assistant Professor, Department of Bioengineering, George R. Brown School of Engineering, Rice University
Previous studies have already established that the neonatal fragment crystallizable receptor (FcRn), a gene involved in regulating the number of antibodies throughout the body, is how antibodies penetrate the blood-brain barrier.
To take advantage of this biological escape hatch, Jerzy Szablowski and colleagues used advanced bioengineering techniques to connect the portion of the antibody that facilitates passage through the blood-brain barrier to a common reporter protein. The researchers were subsequently able to observe that expression mirrored in the mouse's blood when they linked the RMAs to a particular gene and expressed that gene in the mouse's brain.
Jerzy Szablowski said, “This method is very sensitive and can track changes in specific cells; producing this protein in approximately 1% of the brain raised its blood levels up to 100,000-fold compared to baseline. We could specifically track the expression of this one protein with just a blood test.”
For the time being, RMAs are crucial research instruments in Szablowski's opinion, helping scientists better track gene expression in the brain. He mentioned that the RMA platform might be utilized to examine the duration of time that innovative gene therapies remain in the brain.
He explained, “We could track these new therapies with just a blood test and continue to monitor them over time since the RMA platform is non-invasive, but we can also use RMAs to study gene expression as it relates to disease.”
Jerzy Szablowski concluded, “Being able to track different gene expression changes will allow us to understand what leads to disease and how the disease itself changes gene expression in the brain. This could provide new clues for drug development, or even for how to prevent neurological diseases in the first place.”
Source:
Journal reference:
Lee, S., et.al., (2024). Engineered serum markers for non-invasive monitoring of gene expression in the brain. Nature Biotechnology. doi.org/10.1038/s41587-023-02087-x