Caixia Gao’s group at the Chinese Academy of Sciences’ Institute of Genetics and Developmental Biology has created a number of new prime editors based on the Cas12a protein, broadening the applications and targeting range of precision genome editing, as reported in a study published in Nature Biotechnology.
 Prime editors using Cas12a and circular RNAs in human cells. Image Credit: Caixia Gao’s group
Prime editors using Cas12a and circular RNAs in human cells. Image Credit: Caixia Gao’s group
Targeted and accurate DNA insertions, deletions, and replacements are made possible by prime editing (PE). Cas9 has served as the foundation for all effective prime editors to date. However, the Cas9 has some drawbacks that restrict prime editors’ widespread use. For instance, it is too big, more off-target editing occurs, and it performs poorly in areas with a high concentration of G/C.
Solving these issues would increase prime editing’s usefulness and make genome edits more flexible. The Cas12a protein and its crRNA are easier to administer and have a smaller molecular size than Cas9. Additionally, because of its distinct T-rich PAM preference, Cas12a is better able to target A/T-rich regions and has a lower off-target effect. As a result, prime editors built on top of Cas12a will have a variety of possible uses.
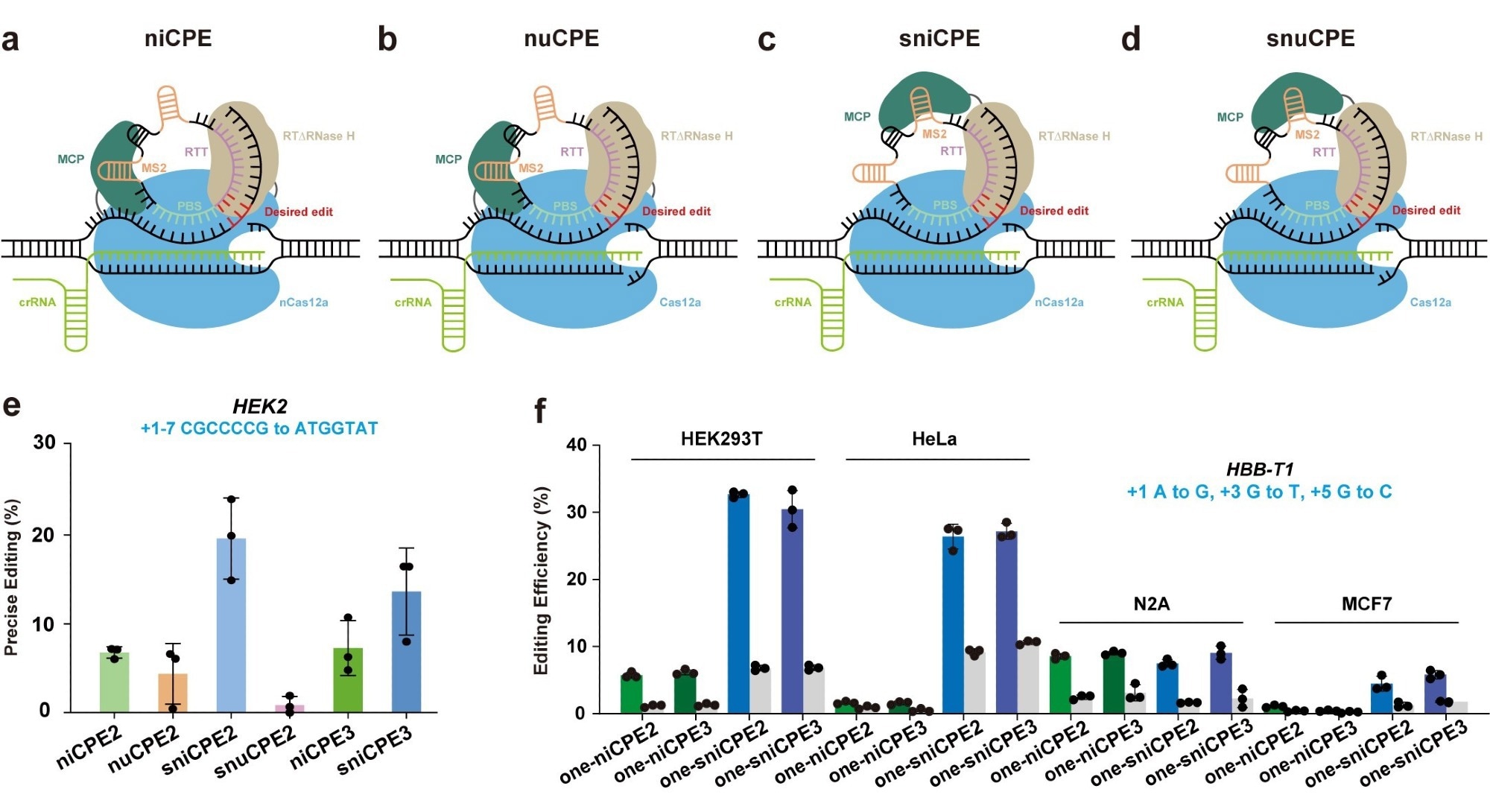
Cas12a protein is known to cleave and process its crRNA, making it unsuitable for use with traditional pegRNAs since the protein cleaves the extensions of the primer binding site (PBS) and reverse transcriptase template (RTT).
The scientists created a suite of distinct Circular RNA-mediated Prime Editors (CPEs) for diverse editing scenarios by utilizing distinct mutant versions of the Cas12a protein. These CPEs include a split nickase-dependent CPE (sniCPE), a nuclease-dependent CPE (nuCPE), a nickase-dependent CPE (niCPE), and a split nuclease-dependent CPE (snuCPE).
The researchers report that in HEK293T cells, the nuCPE and snuCPE achieved high editing efficiencies of up to 10.42% and 3.19%, respectively. Even greater editing efficiencies were attained by the niCPE and sniCPE editors, who scored 24.89% and 40.75%, respectively.
niCPE and sniCPE were also successful in precisely editing HeLa, N2A, and MCF7 cells in addition to HEK293T cells. Importantly, by separating the reverse transcriptases, the sniCPE and snuCPE systems can eventually be delivered using AAV systems.
The researchers demonstrated effective multiplex prime editing by simultaneously expressing all RNAs containing circular RNA under a single promoter. The researchers successfully prime edited two, three, or even four genes using this method.
In the end, the researchers assessed the CPEs’ off-target effects and demonstrated that the researchers did, in fact, have excellent specificity. The newly developed suite of CPE systems with low off-target effects and high editing efficiency provides guidance for the development of future prime editors that employ other nucleases and are diverse in size and targeting capabilities.
These CPE systems will ultimately be very useful for crop breeding, disease treatment, and biological research.
Source:
Journal reference:
Liang, R., et al. (2024) Prime editing using CRISPR-Cas12a and circular RNAs in human cells. Nature Biotechnology. doi.org/10.1038/s41587-023-02095-x