Researchers at the RIKEN Center for Biosystems Dynamics (BDR) in Japan have made a novel discovery that challenges long-held beliefs about DNA replication. The results, led by Ichiro Hiratani and colleagues, were published in Nature on August 28th, 2024. They demonstrate that DNA replication in early embryos differs from what previous studies have shown and involves an unstable phase that is prone to chromosomal copying mistakes.
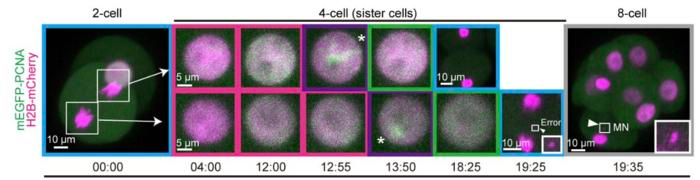 Live imaging of embryos at different stages. Chromosomes are labeled in magenta. 4-cell sister blastomeres are shown, along with 3D-reconstructed images of 2-to-4-cell and 4-to-8-cell divisions. Chromosomal copying errors were most prevalent at this stage of embryogenesis (13% of cells). Image Credit: RIKEN
Live imaging of embryos at different stages. Chromosomes are labeled in magenta. 4-cell sister blastomeres are shown, along with 3D-reconstructed images of 2-to-4-cell and 4-to-8-cell divisions. Chromosomal copying errors were most prevalent at this stage of embryogenesis (13% of cells). Image Credit: RIKEN
The results could have an influence on reproductive medicine by improving the techniques for in vitro fertilization (IVF), as developmental problems and unsuccessful pregnancies are frequently linked to chromosomal abnormalities.
During embryogenesis, both the fertilized egg and each new pair of daughter cells divide. By the third day following fertilization, an embryo has undergone three divisions and has 16 cells. Each cell division is followed by DNA replication, which ensures that each daughter cell has a complete copy of the genome.
In their latest study, a team of RIKEN BDR researchers wanted to describe the nature of DNA replication in early-stage embryos. They utilized their own single-cell genomics technology, known as scRepli-seq, to grow mouse embryos.
Using this approach, the team was able to capture images of single embryonic cell DNA at various stages of replication. What they discovered contradicts what scientists had previously understood about DNA replication in embryos.
We found multiple specialized types of DNA replication during early mouse embryogenesis, which no one has seen before. In addition, we also found that at certain points, genomic DNA is temporarily unstable and chromosomal aberrations are elevated.”
Ichiro Hiratani, PhD, Team Leader, Laboratory for Developmental Epigenetics, RIKEN Center for Biosystems Dynamics Research
According to textbooks, DNA does not replicate in a single step. Instead, certain parts of a chromosome are duplicated in a specified order. The team's initial observation was that replication-timing domains seen in adult cells do not appear until an embryo has four cells. This means that, unlike other cells in the organism, DNA in 1- and 2-cell embryos replicates evenly rather than sequentially.
Each time a chromosome unwinds for replication, sections of DNA unzip, producing a structure like a fork in the road. For replication to occur, the fork must advance down the strand of DNA, rezipping copied sections and unzipping the following part. The team's second observation was that fork speed is substantially slower in the first, second, and fourth cell stages than after the eighth cell stage of embryogenesis.
The 4-cell embryo can now be viewed as a transitional stage in which uniform DNA replication becomes sequential while retaining the sluggish fork movement characteristic of 1- and 2-cell embryos. In contrast, 8-cell embryos behave considerably more like adult cells, with sequential replication and rapid fork movement.
Chromosome abnormalities, such as excess copies, missing copies, breaks in copies, or incomplete copies, are frequently caused by errors in DNA replication that occur in the first few days following fertilization.
While some of these copying errors result in miscarriages, others cause developmental problems such as trisomy 21, often known as Down syndrome. The final finding made by the scientists was that early-stage embryos had a transiently higher incidence of chromosomal copying mistakes, most frequently at the 4-cell stage.
Once more, the researchers employed scRepli-seq to find abnormalities in the copy number of chromosomes. They discovered that extremely few mistakes happened when cells were switching from one to two or from eight to sixteen phases.
However, 13% of cells displayed chromosomal abnormalities as they went from the 4- to the 8-cell stage, most likely as a result of copying mistakes made during the 4-cell stage. Additional testing revealed that the slow-moving forks were connected to the copying problems at this point.
Hiratani added, “Our findings lead to many new questions. For example, are these series of phenomena evolutionarily conserved in other species, including human embryos? And what are the subsequent fates of cells with chromosomal aberrations?”
In addition to directing future fundamental research, this discovery could help fertility clinics develop better procedures for reducing the chromosomal abnormalities that are common in the initial few days following fertilization.
Source:
Journal reference:
Takahashi, S., et al. (2024) Embryonic genome instability upon DNA replication timing program emergence. Nature. doi.org/10.1038/s41586-024-07841-y.