Reviewed by Danielle Ellis, B.Sc.Nov 29 2024
The cell creates mRNA, a copy of DNA, to assemble proteins. The mRNA is then read by a different molecule known as a ribosome, which converts it into protein. However, the process by which the ribosome binds to and reads mRNA was previously unknown to scientists, making this stage a visual enigma.
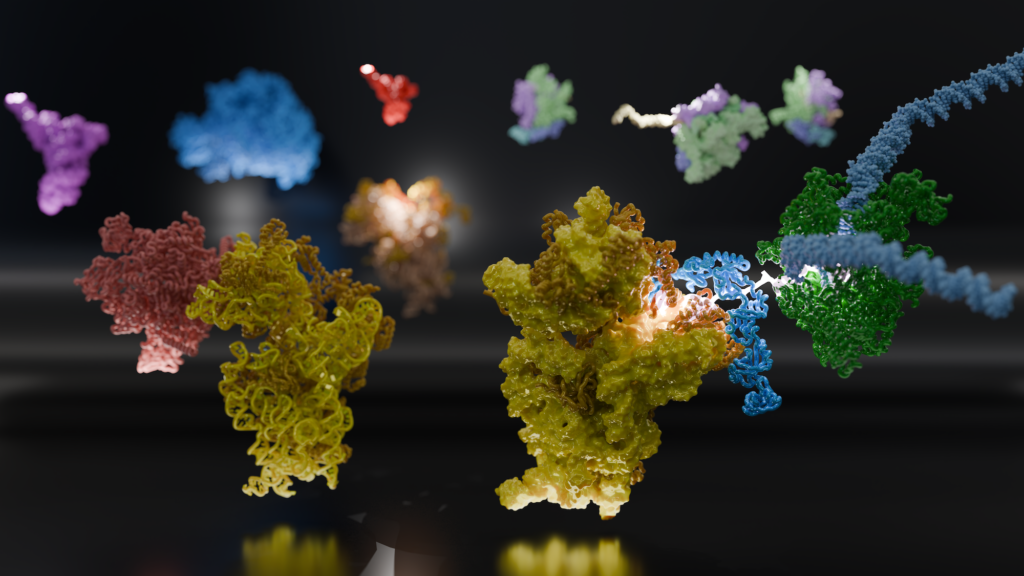 This image highlights two alternatives for the ribosome to be recruited to an mRNA that is still being synthesized by RNA polymerase (RNAP). RNAP (left, red) can directly deliver the mRNA to the entry channel of the small ribosomal subunit (left, yellow). Alternatively, and likely dominant in bacteria, RNAP (green) can interact with ribosomal protein bS1 (right, cyan). bS1 binds and guides the mRNA (right, white, and glowing) from RNAP into the small ribosomal subunit (right, yellow). Image Credit: Mohammad Afsar and Huma Rahil
This image highlights two alternatives for the ribosome to be recruited to an mRNA that is still being synthesized by RNA polymerase (RNAP). RNAP (left, red) can directly deliver the mRNA to the entry channel of the small ribosomal subunit (left, yellow). Alternatively, and likely dominant in bacteria, RNAP (green) can interact with ribosomal protein bS1 (right, cyan). bS1 binds and guides the mRNA (right, white, and glowing) from RNAP into the small ribosomal subunit (right, yellow). Image Credit: Mohammad Afsar and Huma Rahil
A group of researchers from around the world, including those from the University of Michigan, have now employed sophisticated microscopy to visualize the recruitment of ribosomes to mRNA during its transcription by the enzyme RNA polymerase, or RNAP. Their study, which looked at the mechanism in bacteria, was published in the journal Science.
Understanding how the ribosome captures or ‘recruits’ the mRNA is a prerequisite for everything that comes after, such as understanding how it can even begin to interpret the information encoded in the mRNA. It is like a book. Your task is to read and interpret a book, but you do not know where to get the book from. How is the book delivered to the reader?”
Albert Weixlbaumer, Researcher and Study Co-Lead Author, Institute of Genetics and Molecular and Cellular Biology, University of Strasbourg
The researchers found that the RNAP that transcribes the mRNA uses two distinct anchors to rope in the ribosome and guarantee a firm footing for the initiation of protein synthesis. This is comparable to a construction site foreperson supervising workers while they install a complicated superstructure part, doing two redundant checks to ensure that every component is fastened securely at crucial points for optimal stability and functionality.
The researchers believe that by comprehending these basic mechanisms, new antibiotics that target these particular pathways in bacterial protein synthesis could be developed. Antibiotics have traditionally targeted the ribosome or RNAP, although bacteria frequently change and mutate to develop some resistance to such drugs. With their newfound understanding, the team intends to outsmart bacteria by blocking several of their paths.
We know there is an interaction between the RNAP, the ribosome, transcription factors, proteins, and mRNA. We could target this interface, specifically between the RNAP, ribosome, and mRNA, with a compound that interferes with the recruitment or the stability of the complex.”
Adrien Chauvier, Study Co-Lead Author and Senior Scientist, University of Michigan
To demonstrate how the many parts of the complex cooperate to deliver newly generated mRNAs to the ribosome and serve as linkages between transcription and translation, the team created a mechanistic framework.
We wanted to find out how the coupling of RNAP and the ribosome is established in the first place. Using purified components, we reassembled the complex 10-billionth of a meter in diameter. We saw them in action using cryo-electron microscopy (cryo-EM) and interpreted what they were doing. We then needed to see if the behavior of our purified components could be recapitulated in different experimental systems.”
Albert Weixlbaumer, Researcher and Study Co-Lead Author, Institute of Genetics and Molecular and Cellular Biology, University of Strasbourg
The walled-off nucleus of more complicated human cells contains DNA, and RNAP acts as the “interpreter,” dissecting genetic instructions into smaller pieces. A carefully chosen copy of a small portion of the genetic code is transferred to the ribosome in the much “roomier” cytoplasm, where it is translated into proteins, the fundamental building blocks of life, by this dynamo of enzymes that transcribe or write, the DNA into mRNA.
In prokaryotes, which lack a distinct nucleus and internal membrane boundaries, transcription and translation occur simultaneously and in close proximity. This enables the RNA polymerase (RNAP) and the ribosome to coordinate their functions and work together efficiently directly.
Bacteria are the most well-understood prokaryotes, and due to their simple genetic structure, they provided the team with an ideal host for analyzing the mechanisms and machinery involved in ribosome-RNAP coupling during gene expression.
To investigate the processes involved, the researchers used a variety of technologies and procedures specific to each lab's area of expertise, such as Weixlbaumer's group's cryo-EM and Andrea Graziadei's group's in-cell crosslinking mass spectrometry.
Chauvier, a Biophysics Expert, and Nils Walter, a Professor of Chemistry, Biophysics, and Biological Chemistry at the University of Michigan, used their sophisticated single-molecule fluorescence microscopes to examine the structure's dynamics.
Chauvier said, “In order to track the speed of this machinery at work, we tagged each of the two components with a different color. We used one fluorescent color for the nascent RNA and another one for the ribosome. This allowed us to view their kinetics separately under the high-powered microscope.”
They found that when ribosomal protein bS1 was present, the mRNA that emerged from RNAP was more effectively attached to the small ribosomal subunit (30S), which aids in the mRNA's unfolding in preparation for translation inside the ribosome.
By tethering RNA polymerase with the coupling transcription factor NusG or its paralog or version, RfaH, which threads the mRNA into the ribosome's mRNA entry channel from the opposite side of bS1, Webster and Weixlbaumer's cryo-EM structures identified an alternate route of mRNA delivery to the ribosome.
The team anticipates more cooperation to determine how the complex must reorganize to become fully functioning after successfully visualizing the initial step of establishing the link between RNAP and the ribosome.
Source:
Journal reference:
Webster, M. W., et al. (2024) Molecular basis of mRNA delivery to the bacterial ribosome. Science. doi.org/10.1126/science.ado8476.