Biochemists at the National University of Singapore (NUS) have identified a new subclass of trifunctional enzymes in gram-positive bacteria that play an important role in the manufacture of an antibiotic lanthipeptide.
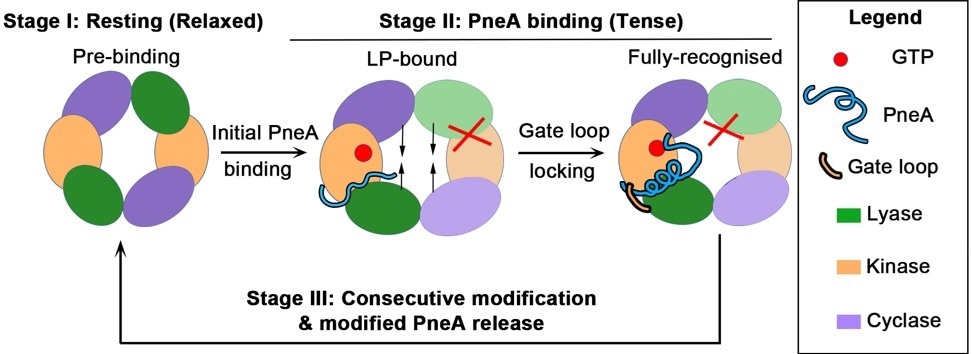 Schematic illustrates the initial stages of substrate PneA binding by enzyme PneKC, which shows the relaxed prebinding state, followed by overall tensing induced by substrate binding to one protomer. This conformational change likely prevents substrate binding to the other protomer, providing a basis for the negative cooperation in enzymatic activity observed. Image Credit: Nature Communications
Schematic illustrates the initial stages of substrate PneA binding by enzyme PneKC, which shows the relaxed prebinding state, followed by overall tensing induced by substrate binding to one protomer. This conformational change likely prevents substrate binding to the other protomer, providing a basis for the negative cooperation in enzymatic activity observed. Image Credit: Nature Communications
Lanthipeptides are a type of ribosomally synthesized and post-translationally modified peptide (RiPP) produced by gram-positive bacteria in response to environmental stress. They are essential biomolecules with antibacterial, antifungal, and antiviral properties that show promise for developing new drugs.
Despite their therapeutic importance, the mechanism behind the post-translational modification of a particularly potent family of these peptides is largely unknown.
The study team led by Assistant Professor Min Luo from the Department of Biological Sciences at NUS characterized a unique subclass of lanthipeptide modifying enzymes using a combination of bioinformatics, structural characterization using cryo-electron microscopy (cryoEM), and functional testing.
These enzymes are in charge of identifying the lanthipeptide precursor and progressively introducing several types of post-translational modifications, which transform the peptide into its functional form.
They identified the first homodimeric lanthipeptide modifying enzyme, PneKC, from the bacterium Streptococcus Pneumonae. Structural research of the enzyme identified a key area, or “dimerization hotspot,” where two components of the enzyme combine to create a dimer. When this area was altered, the enzyme lost its ability to form the dimeric structure.
The research was published in the journal Nature Communications.
Functional experiments revealed that the enzyme dimer regulates itself through a process known as negative cooperativity. When a substrate binds to one-half (protomer) of a dimer, it causes it to shift shape. This modification is passed across the dimerization interface to the unbound protomer, influencing its activity.
The researchers also discovered the early steps of peptide recognition and hypothesized a set of previously unknown catalytic residues, which are specific amino acids within the enzyme’s active site that aid in the last phase of peptide modification. These catalytic residues are critical in driving the cyclization phase, which allows the peptide to mature into its active form as lanthipeptide.
This phase had long been a mystery in this enzyme class, and the discovery of these catalytic residues adds a vital piece to the puzzle, improving the understanding of how these enzymes facilitate the complicated changes required for lanthipeptide production.
Following this discovery, Ms Yifan LI, a PhD candidate on the research team and the first author of this study, is now attempting to identify the following stages of lanthipeptide alteration.
Her current study attempts to better understand the maturation processes and pharmacodynamics of these recently discovered lanthipeptides, with the goal of enhancing their therapeutic applications.
Our current study significantly enhanced our understanding of the dynamic processes underlying peptide recognition and domain coordination within lanthipeptide modification enzymes. Moreover, considering the high potential of lanthipeptides in antimicrobial applications, these insights pave the way for the development of lanthipeptides for antimicrobial purposes.”
Min Luo, Assistant Professor, Department of Biological Sciences, National University of Singapore
Source:
Journal reference:
Li, Y., et al. (2024) Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers. Nature Communications. doi.org/10.1038/s41467-024-51600-6.