Fluorescence microscopy has allowed scientists to overcome the lesser resolving power of ordinary optical microscopes using carefully designed fluorophore tags. However, the method is not without its limitations. This article will briefly describe what has made fluorescence microscopy such a popular analytical tool in the biosciences, and some of the limitations encountered so far.
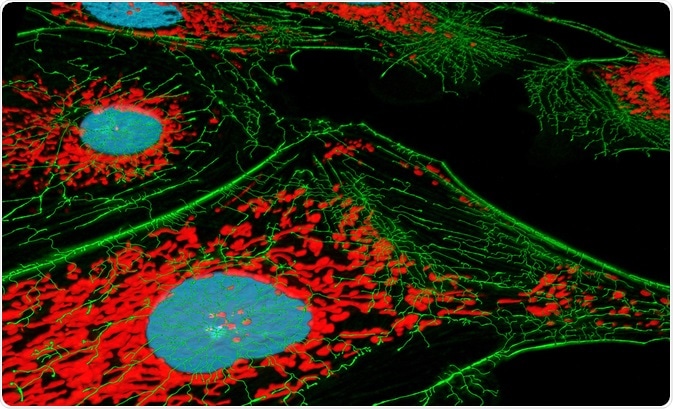
Image Credit: Heiti Paves/Shutterstock.com
What are the Advantages?
Fluorescence microscopy is among the most popular methods of live-cell observation and the structure elucidation of biomolecules in tissues and cells, allowing them to be studied in situ without the need for toxic and time-consuming staining processes. Samples may be fixed before the addition of a fluorophore, halting the metabolism of cells at a point in time and allowing detailed measurements to be made of the preserved sample, or cell dynamics can be examined in living samples with a precision and sensitivity capable of tracking the path of a single protein throughout its lifetime.
The high degree of specificity possible thanks to modern fluorophore probes allows specific proteins or other biological structures to be emphasized and highlighted, providing a clear three-dimensional internal structure of the cell when used in combination with confocal microscopy. The diversity and specificity of the available probes allow multiple overlapping structures to be identified simultaneously within the same sample.
For example, commonly employed probe 4′,6-diamidino-2-phenylin-dole (DAPI) forms a fluorescent complex following binding with DNA, appearing blue, while another probe branded MitoTracker Red interacts with mitochondria, then appearing red. Both of these probes can be used simultaneously to identify the structure and distribution of both the nucleus and mitochondria in living cells.
The sensitivity of these probes is great enough to distinguish between these features clearly, only requiring a few dozen active fluorophores in a cubic micrometer of space to produce a detectable signal, with this figure improving over time as improved methods of contrast enhancement are implemented.
What are the Limitations?
Despite the diverse range of probes available, fluorescence microscopy is limited by its dependence on probes. Molecular structures that are unlabeled and incapable of autofluorescence will remain dark in the final image, and thus unexpected or novel structures cannot be observed without initial identification and careful probe design or selection.
Additionally, the simultaneous use of multiple probes or the presence of autofluorescent biomolecules may induce interactions that detract from the effectiveness of each probe, causing a poorer signal-to-noise ratio. Many molecules found natively within cells, and even other fluorophores, are capable of quenching a fluorescent signal.
Quenching can occur in several ways, all of which necessitate close contact between the fluorophore and quencher. Interestingly, this phenomenon has been employed to signal the release of a fluorophore from an intentionally bound quencher, with practical applications including the indication of the presence of an enzyme that it was known could cleave the bond.
The cells under investigation may themselves be damaged by the process of fluorescence microscopy, from both exposure to light and the fluorophores themselves. The lower wavelengths of light used to excite the fluorophore are particularly damaging to cells, both directly damaging DNA and producing reactive oxygen species that go on to affect cell viability.
In many cases, this is not an issue, as the desired information can be collected quickly enough that cell damage is not observed. However, in time-course studies that depend on a repeated examination of the same cells, the cumulative effect can be significant, not only impacting cell viability but causing increased cell membrane permeability and degradation of the cytoskeleton, potentially negatively impacting any data gathered.
Photobleaching is an issue inherent among all small molecule and protein fluorophores, leaving them completely without fluorescence. Photobleaching may occur through several mechanisms that are still not completely elucidated. Typically, upon excitation, a fluorophore may promote an electron to a higher energy level in either a singlet or triplet state, meaning that the electron either maintains or reverses its orientation, respectively.
The triplet state is both more reactive and long-lived than its singlet counterpart, as it is difficult to return to the prior energy level already occupied by an electron of the same spin due to the Pauli exclusion principle, and an excited fluorophore may interact with neighboring molecules to cleave or form covalent bonds.
Tissue fixation requires several fixing and washing steps that can be both time-consuming and omit structural details of the cell. Formaldehyde is frequently used for this purpose and fixes the structural orientation of proteins by cross-linking. However, lipids and small molecules within the cell are often lost in this process.
Imaging live cells impose several issues regarding maintaining cell viability throughout the observation period. Besides the damaging effects of photon bombardment to the cells and fluorophores described above, the cell culture plate must be kept in suitable atmospheric conditions. The temperature needs to remain constant without significant fluctuations, both for the health and normal function of the cell and because even minor changes can alter the refractive index of the material, altering the focal point of the laser and microscope.
The atmospheric concentration of CO2 and water vapor must also be carefully controlled. CO2 is closely correlated with the pH of the culture media, and many fluorophores are pH dependant, being strongly quenched outside of their operating range. Low atmospheric humidity encourages media evaporation, altering the volume of media and therefore the concentration of compounds within the media over time.
Sources:
- Kapuscinski, J. (1995) DAPI: a DNA-specific Fluorescent Probe. Biotechnic & Histochemistry, 70(5), pp.220-233. https://www.tandfonline.com/doi/abs/10.3109/10520299509108199
- Zhuang, X., Ha, T., Kim, H. D., Center, T., Labeit, S. & Chu, S. (2000) Fluorescence quenching: A tool for single-molecule protein-folding study. PNAS, 97(26), pp.14241-14244. https://www.pnas.org/content/97/26/14241
- Simeonov, A. & Davis, M. I. (2015) Interference with Fluorescence and Absorbance. In: Markossian S., Sittampalam G. S., Grossman A., et al., editors. Assay Guidance Manual [Internet]. Bethesda (MD): Eli Lilly & Company and the National Center for Advancing Translational Sciences https://www.ncbi.nlm.nih.gov/books/NBK343429/
- Wäldchen, S., Lehmann, J., Klein, T., van de Linde, S. & Sauer, M. (2015) Light-induced cell damage in live-cell super-resolution microscopy. Scientific Reports, 5. doi:10.1038/srep15348 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4611486/
- chem.libretexts.org/.../14.7%3A_Fluorescence_and_Phosphorescence
- Smith, C. L. (2011). Basic Confocal Microscopy. Current Protocols in Neuroscience. doi: 10.1002/0471142301.ns0202s56 currentprotocols.onlinelibrary.wiley.com/.../0471142727.mb1411s81
Further Reading
Last Updated: Apr 2, 2025