Single-cell proteomics, which involves identifying and characterizing proteins within single cells, can be performed using techniques such as mass cytometry and ion beam imaging. These assays can provide insight into complex biological systems, enhance our understanding of cellular heterogeneity, and ultimately help to improve patient outcomes.
Image Credit: Design_Cells/Shutterstock.com
Single-Cell Proteomics
Proteins are essential to a wide range of cellular functions and processes. This includes regulating gene expression, catalyzing biochemical reactions, and transporting molecules. As such, protein characterization provides a vast amount of information on biological systems.
However, traditional methods of protein measurement such as western blotting, mass spectrometry, and enzyme-linked immunosorbent assays are population-based approaches that can mask functional or morphological differences between cells, known as cellular heterogeneity.
Non-genetic cellular heterogeneity is integral to processes such as stem cell differentiation, cancer metastasis, and immune responses. For instance, Ma et al. (2011) found profound differences in the cytokine secretion profiles of T-cell populations previously considered to be homogenous. This functional diversity was more profound amongst cell populations that were responding to tumors compared to those that were obtained from healthy donors.
Characterizing cellular heterogeneity is, therefore, critical for furthering our understanding of normal cell activity and disease processes. This can, in part, be achieved using single-cell proteomic technologies, which aim to catalog and characterize the total complement of protein isoforms in single cells.
How is Single-Cell Proteomic Analysis Conducted?
Because of the limited amount of protein in individual cells, single-cell protein analysis must be conducted using highly sensitive techniques that allow multiple proteins to be analyzed simultaneously.
Single-cell proteomic analysis is often carried out on isolated cells using techniques such as mass cytometry. During mass cytometry, single-cell protein targets are stained with antibodies conjugated to different metal isotopes. Individual cells are then sprayed as single droplets and subsequently ionized.
A time-of-flight mass spectrometer quantifies the ions generated from each cell, and the metal isotopes' signals are translated into the expression levels for each protein target. Mass cytometry facilitates the analysis of approximately 1000 cells per second, giving it the highest sample throughput of all current single-cell proteomic techniques.
Protein analysis in isolated single cells can also be conducted using single-cell barcode chips, which enclose cells in chambers along with an array of immobilized antibody strips. After lysis or secretion, proteins bind to the immobilized antibodies and are detected using a second fluorescent antibody. The precise location of the fluorescence is used to identify the target protein.
Most single-cell proteomic assays use antibodies to detect proteins, which can lead to problems with cross-reactivity and non-specific binding, thereby generating false positive signals and decreasing accuracy. Single-cell Western blots can overcome these problems, as proteins are separated by mass before antibody labeling.
Single-cell Westerns use a thin layer of polyacrylamide gel containing thousands of microwells to trap individual cells. Cells are lysed, and proteins within the gel are separated based on molecular mass. The separated proteins can then be detected using fluorescent antibodies.
Although single-cell Western blots allow over 6,000 cells to be analyzed at once, this technique can suffer from limited detection sensitivity, as a substantial proportion of protein is lost during the assay.
Single-cell protein analysis can also be conducted using in situ techniques, such as imaging mass cytometry and ion beam imaging. These methods allow protein profiling to be carried out while cells remain in their natural spatial contexts. This helps to better understand how the location of cells within a tissue can affect the function and regulation of biological systems.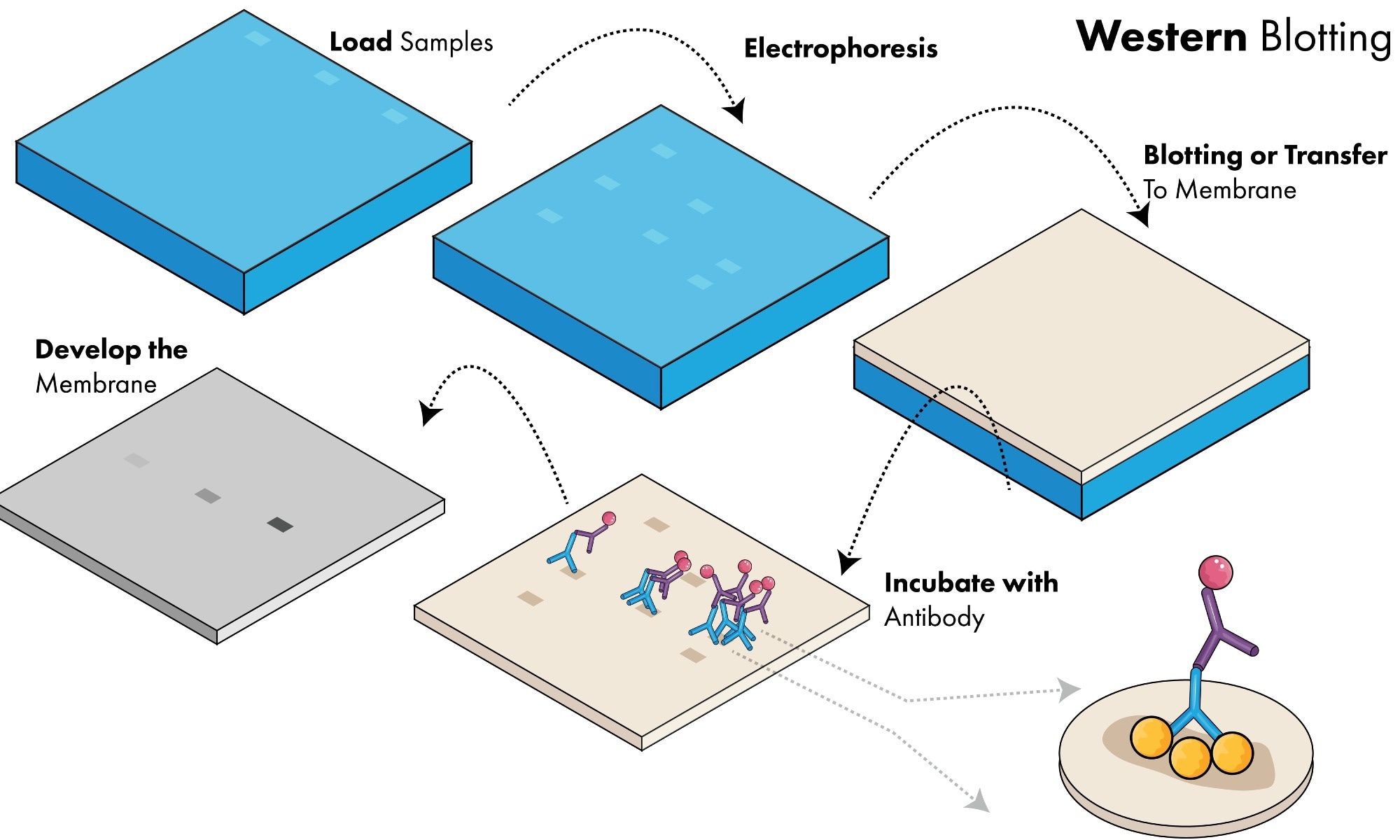
Image Credit: Art of Science/Shutterstock.com
Applications and Future Directions
Single-cell proteomics has a wide range of biological applications. For instance, mass cytometry has been used to investigate immune cell heterogeneity, and single-cell Western blots have enhanced our understanding of cancer cell heterogeneity.
Cell-cell interactions and tissue organization have been probed through in situ proteomic analysis of intact brain and tumor tissues, and single-cell barcode chips have been used to study signaling pathways in cancer cells and immune cells. This investigation of intracellular signaling networks could lead to new discoveries on protein function, biological responses to drugs, and mechanisms of drug resistance.
Single-cell proteomics may also lead to the discovery of new protein biomarkers, as it allows protein abundance and location to be compared between individual cells in normal and diseased tissues. These new biomarkers could facilitate more effective disease diagnosis and improve the monitoring of immune responses to treatment.
However, despite the insight these techniques provide and the biomedical applications of these insights, single-cell proteomic assays are not without their issues. Currently, their limited multiplexing capacity is a significant drawback, as this means only a relatively small fraction of proteins can be studied at one time.
If single-cell proteomic techniques were combined with other assays, it may help to overcome this problem. For example, if alternative approaches were used to identify the different cell subtypes present within a sample, along with their active pathways, researchers could choose the most informative proteins to profile using single-cell proteomics.
Sources:
- Pham, T., Tyagi, A., Wang, Y.S. and Guo, J. (2021). Single‐cell proteomic analysis. WIREs Mechanisms of Disease, 13(1). https://doi.org/10.1002/wsbm.1503
- Ma, C., Fan, R., Ahmad, H., Shi, Q., Comin-Anduix, B., Chodon, T., Koya, R.C., Liu, C.C., Kwong, G.A., Radu, C.G. and Ribas, A. (2011). A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nature Medicine, 17(6), pp.738-743. https://doi.org/10.1038/nm.2375
- Vistain, L.F. and Tay, S. (2021). Single-cell proteomics. Trends in Biochemical Sciences, 46(8), pp.661-672.
- Khare, K. and Pandey, R., 2022. Cellular heterogeneity in disease severity and clinical outcome: Granular understanding of immune response is key. Frontiers in Immunology, 13. https://doi.org/10.3389/fimmu.2022.973070
Further Reading
Last Updated: May 3, 2023