A research team from China has now created two novel compounds that block the SARS-CoV-2 main protease (Mpro). One of the compounds is an ideal drug candidate for further clinical studies.
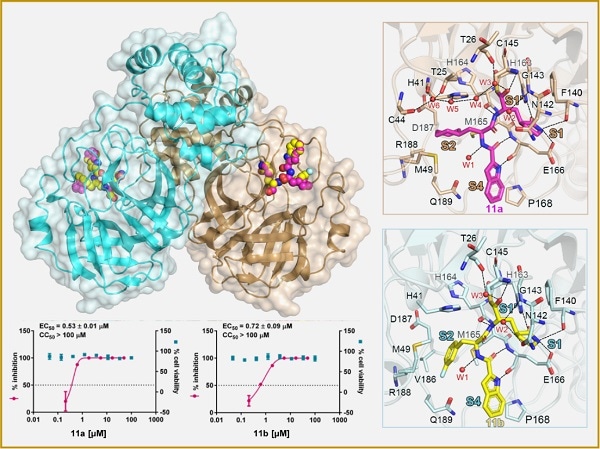
Mpro-inhibitor binding modes for 11a and 11b. Image Credit: SIMM.
The study was performed by Prof. LIU Hong and Prof. XU Yechun from the Shanghai Institute of Materia Medica (SIMM) of the Chinese Academy of Sciences (CAS), Prof. YANG Haitao from the Shanghai Institute for Advanced Immunochemical Studies of ShanghaiTech University, Professor ZHANG Lei-Ke from the Wuhan Institute of Virology of CAS, and their colleagues. It was published online in Science on April 22nd.
Until April 22, there have been over 2.5 million confirmed COVID-19 cases, accounting for over 170,000 mortalities. At present, there are no particular antiviral drugs or clinically effective vaccines for the prevention and treatment of COVID-19.
SARS-CoV-2 is the causative agent responsible for the global COVID-19 outbreak. It consists of a positive-sense, single-stranded RNA virus protected by an envelope, and SARS-CoV-2 Mpro plays a significant role in its life cycle. SARS-CoV-2 Mpro was found to be an ideal antiviral drug target as it does not have a human homologue.
Researchers designed and produced two novel compounds called 11a and 11b, after the analysis of substrate-binding pockets of SARS-CoV-2 Mpro. Then, a fluorescence resonance energy transfer (FRET)-based cleavage assay was employed to identify their IC50 values.
The outcomes showed excellent SARS-CoV-2 Mpro inhibitory action for both 11a and 11b, with IC50 values of 0.053 ± 0.005 μM and 0.040 ± 0.002 μM, respectively.
The researchers also made use of quantitative real-time PCR, immunofluorescence, and plaque assay to track the antiviral activity of 11a and 11b.
The findings revealed that the compounds 11a and 11b showed good anti-SARS-CoV-2-infection activity in cell culture—for example, EC50 values were 0.53 ± 0.01 μM and 0.72 ± 0.09 μM, respectively, when the plaque assay was used.
Moreover, these compounds exhibited good PK properties in vivo, which suggested that they were the potential drug candidates. But the minimal toxicity of the compound 11a made it favorable.
Researchers established a high-resolution crystal structure of complexes Mpro-11a (PDB: 6LZE) and Mpro-11b (PDB: 6M0K) at 1.5-A resolution for finding the blocking mechanism of SARS-CoV-2 Mpro in compounds 11a and 11b.
This high-resolution crystal structure of the complexes revealed the mechanism of SARS-CoV-2 inhibition besides depicting the interactions among SARS-CoV-2 Mpro-11a/11b. High-resolution analysis of complexes helped medicinal chemists to design novel inhibitors that act against SARS-CoV-2.
This study reveals that the structure-based drug design is an efficacious approach to design particular antiviral lead compounds against SARS-CoV-2. Preclinical studies on compound 11a are underway. The researchers intend to share the research data with researchers worldwide to expedite the development of anti-SARS-CoV-2 drugs.
The group led by Professors LIU Hong, XU Yechun, and JIANG Hualiang, the team achieved the design and production of compounds and determined the inhibitory action of the enzyme.
Another group led by Professors YANG Haitao and RAO Zihe identified the high-resolution crystal structure. A third group led by Professors ZHANG Lei-Ke and XIAO Gengfu investigated the antiviral activity.
The study was financially supported by the National Chengdu Center for Safety Evaluation of Drugs and Frontier Biotechnologies Inc.
Source:
Journal reference:
Dai, W., et al. (2020) Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. doi.org/10.1126/science.abb4489.