About 350 years ago, Robert Hooke provided an initial description of a cell in Micrographia, and since then, microscopy has played a major role in figuring out life’s rules.
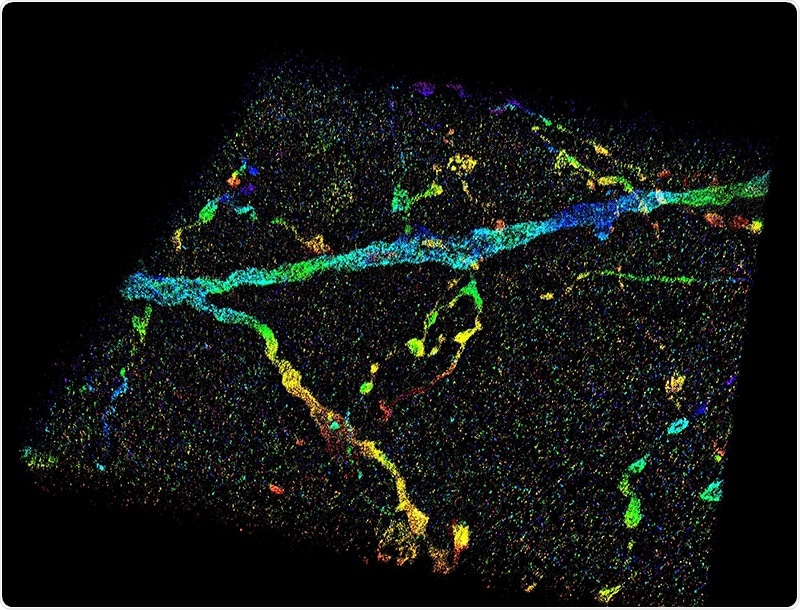
This image shows a 3D super-resolution reconstruction of dendrites in the primary visual cortex. Image Credit: Fang Huang/Purdue University.
Resolution is known to be the smallest resolvable feature and has been limited by the wave nature of light. This barrier had remained for 100 years, limiting one’s interpretation of the dynamics, functions, and interactions of cells, specifically at the sub-micron to the nanometer scale.
This underlying limitation has been resolved by super-resolution fluorescence microscopy, which offers up to 10 times better resolution. This technology enables researchers to observe the internal function of both biomolecules and cells at an unmatched spatial resolution.
But this resolving potential is often obstructed when visualizing the internal parts of whole-tissue or whole-cell specimens, for example, the ones that are usually examined during the investigations of the brain or cancer.
Light signals, produced by molecules within a specimen, move via different portions of the tissue or cell structures at varying speeds and lead to aberrations, which consequently impact the resolutions of the image.
Now, scientists from Purdue University have devised an innovative technology to resolve this problem.
Our technology allows us to measure wavefront distortions induced by the specimen, either a cell or a tissue, directly from the signals generated by single molecules—tiny light sources attached to the cellular structures of interest.”
Fang Huang, Assistant Professor of Biomedical Engineering, College of Engineering. Purdue University
Huang elaborated, “By knowing the distortion induced, we can pinpoint the positions of individual molecules at high precision and accuracy. We obtain thousands to millions of coordinates of individual molecules within a cell or tissue volume and use these coordinates to reveal the nanoscale architectures of specimen constituents.”
The new technology developed by the Purdue research team was recently published in the Nature Methods journal.
“During three-dimensional super-resolution imaging, we record thousands to millions of emission patterns of single fluorescent molecules,” stared Fan Xu, the study’s co-first author and a postdoctoral associate in Huang’s laboratory.
“These emission patterns can be regarded as random observations at various axial positions sampled from the underlying 3D point-spread function describing the shapes of these emission patterns at different depths, which we aim to retrieve.
Xu further added, “Our technology uses two steps: assignment and update, to iteratively retrieve the wavefront distortion and the 3D responses from the recorded single-molecule dataset containing emission patterns of molecules at arbitrary locations.”
The innovative technology devised by the Purdue team may help to detect the locations of biomolecules with accuracy as low as a few nanometers within the whole tissues and cells, and thus overcome the tissue and cellular architectures with a high level of excellent fidelity and resolution.
This advancement expands the routine applicability of super-resolution microscopy from selected cellular targets near coverslips to intra- and extra-cellular targets deep inside tissues. This newfound capacity of visualization could allow for better understanding for neurodegenerative diseases such as Alzheimer’s, and many other diseases affecting the brain and various parts inside the body.”
Donghan Ma, Study Co-First Author and Postdoctoral Researcher, Purdue University
Donghan Ma currently works in Huang’s laboratory. The research received significant support from the National Institutes of Health.
The research team also includes Gary Landreth, a professor in the School of Medicine at Indiana University; Sarah Calve, an associate professor of biomedical engineering in the College of Engineering at Purdue University (presently an associate professor of mechanical engineering at the University of Colorado Boulder); Peng Yin, a Harvard Medical School professor; and Alexander Chubykin, an assistant professor of biological sciences at Purdue University. The entire list of authors is provided in the Nature Methods journal.
This technical advancement is startling and will fundamentally change the precision with which we evaluate the pathological features of Alzheimer’s disease. We are able to see smaller and smaller objects and their interactions with each other, which helps reveal structure complexities we have not appreciated before.”
Gary Landreth, Professor, School of Medicine, Indiana University
According to Calve, this novel technology represents a major innovation towards regenerative treatments to help support the repair mechanisms inside the body.
“This development is critical for understanding tissue biology and being able to visualize structural changes,” stated Calve.
According to Chubykin, whose laboratory investigates autism and other disorders that affect the brain, the high-resolution imaging technology offers a new approach for figuring out the brain’s impairments.
“This is a tremendous breakthrough in terms of functional and structural analyses,” stated Chubykin. “We can see a much more detailed view of the brain and even mark specific neurons with genetic tools for further study.”
The scientists have patented the technology in collaboration with the Purdue Research Foundation Office of Technology Commercialization.
The office has currently shifted to the Convergence Center for Innovation and Collaboration in Discovery Park District, beside Purdue University’s campus. The researchers are now seeking partners to commercialize their novel imaging technology.
Source:
Journal reference:
Xu, F., et al. (2020) Three-dimensional nanoscopy of whole cells and tissues with in situ point spread function retrieval. Nature Methods. doi.org/10.1038/s41592-020-0816-x.