At the University of Groningen, researchers have devised a new technique that integrates different levels of resolution in a computer simulation of biological membranes.
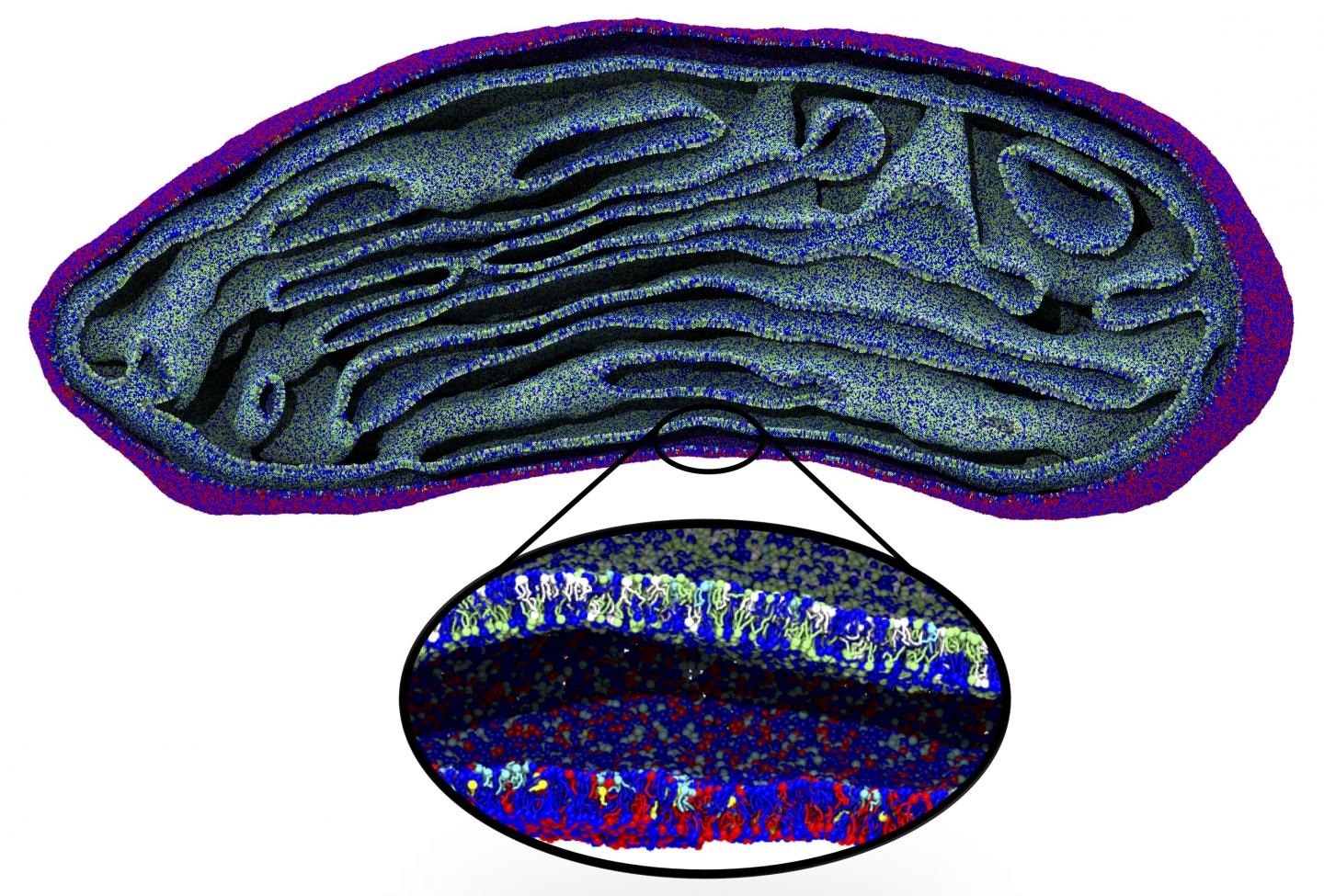
Snapshot of the membranes of an entire organelle, a mitochondrion, simulated for the first time at molecular resolution. Image Credit: Marrink Lab, University of Groningen.
The researchers’ new algorithm back maps a massive model that comprises features like membrane curvature to its matching coarse-grained molecular model.
This has enabled them to focus on toxin-induced membrane budding and to replicate a full-sized mitochondrial lipid membrane.
Published in the Nature Communications journal on May 8th, 2020, the researchers’ method paves the way to whole-cell simulations at a molecular scale.
Molecular dynamics simulations are a robust tool to analyze the interactions and movements of molecules and atoms. But in several biological processes, major changes in, for instance, the shape of the membrane are crucial.
These shape changes are of fundamental importance to the cells functioning, however, the time and length scale of these membrane shape changes are too large for simulations at a molecular resolution.”
Siewert-Jan Marrink, Professor of Molecular Dynamics, University of Groningen.
Challenging
Although increased computing power enables longer and more intricate simulations, cell structures like mitochondria are still out of reach. As a result, the Molecular Dynamics group has created an algorithm that connects massive changes to the simulations at the molecular level.
The group began with an electron micrograph density map in the case of mitochondria.
The densities were converted into lipid structures that were subsequently applied as the input for a molecular dynamics simulation with the Coarse Grain (CG) Martini force field, which was earlier created by Marrink.
The difficult part is to place the lipids in the correct orientation in this density map, which is especially challenging in bent areas.”
Wria Pezeshkian, Study Co-Author, University of Groningen
Pezeshkian is also a postdoctoral researcher in Marrink’s team.
The new algorithm enables users to include varying kinds of lipids to the membrane, at a sensible packing level. Marrink and his collaborators used this approach to replicate the whole lipid membrane of a mitochondrion for 2 ns.
“This structure contained more than five million lipids, which meant that the simulation had to deal with 80 million particles as each lipid molecule consists of multiple particles,” added Pezeshkian.
Triangles
Given the shape and size, the complexity of this simulation is greater than any simulation carried out before.
A simulation of microseconds would have been possible but, as we had no information on the localization of the proteins in the mitochondrial membrane, it only contained lipids and is therefore unstable.”
Siewert-Jan Marrink, Professor of Molecular Dynamics, University of Groningen
It is indeed possible to add this additional complexity to the simulation, and this is presently ongoing.
Rather than a destiny map, the input meant for the system could even be a continuum model, which denotes the surface of the membrane as triangles composed of nodes that are linked by “'springs.” A model like this can estimate forces produced by the deformation of the membrane.
Backmapping toxin proteins and lipids onto the matching parts of this model enabled Marrink and his collaborators to gain in-depth insights into the molecular behavior in the stalk of a membrane bud caused by the joint action of several toxins.
Synthetic cell
Marrink added, “Our final goal is to simulate an entire eukaryotic cell and zoom in on specific parts of this object.”
At present, this is out of reach, albeit the present system already enables the replication of massive objects within a cell, for example, the Golgi apparatus or the endoplasmic reticulum. “And we could probably simulate a red blood cell,” added Marrink.
A basic synthetic cell may soon be accessible. Marrink is engaged in a project that aims to produce a synthetic cell and being able to replicate processes like cell division would assist its design.
“We would really like to know which lipids and proteins could play a role in cell constriction during division,” Marrink concluded.
Source:
Journal reference:
Pezeshkian, W., et al. (2020) Backmapping triangulated surfaces to coarse-grained membrane models. Nature Communications. doi.org/10.1038/s41467-020-16094-y.