Scientists can gain a better understanding of the cellular roles of the disrupted gene by disturbing the expression of a specific gene and tracking how this change has an impact on the expression of other types of genes.
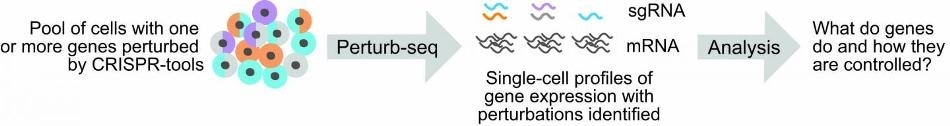
Improvements by the team allow scientists to more easily link genetic perturbations to changes in single-cell gene expression profiles, and to perform Perturb-seq studies more cheaply. Image Credit: Replogle, et al.
Although innovative technologies, like Perturb-seq, provide unparalleled detail and deeper insight from those genetic disruption analyses, practical and technical barriers have restricted the use of Perturb-seq technology.
New research, performed by Princeton University researcher Britt Adamson and his collaborators from biotech company 10X Genomics and the University of California-San Francisco, aims to change all that. The study was published in the Nature Biotechnology journal on March 30th, 2020.
We present several improvements to this approach, which together laid the groundwork for performing Perturb-seq screens at larger scale and with combinatorial perturbations.”
Britt Adamson, Researcher, Princeton University
The initial step in the Perturb-seq method is to disturb a group of target genes. This is achieved by applying one of a suite of CRISPR-based technologies to edit the genome of a cell.
CRISPR techniques have been adapted from a defense system present in archaea and bacteria. These microorganisms contain DNA stretches—called “clustered regularly interspaced palindromic repeats,” or CRISPRs for short—that were acquired from the viruses that infect them.
CRISPR sequences are used by both bacteria and archaea to create RNA fragments that direct the CRISPR-related (CAS) enzymes to viral genomes (for example, DNA or, in certain cases, RNA). Once these enzymes reach there, they divide the bound genome and stop the infection.
By using synthetic RNAs known as “guides,” or sgRNAs, the researchers were able to reprocess these systems for use in animal cells. These sgRNAs can target the Cas enzymes to the DNA of a cell. At this point, cleavage disrupts the targeted genes by introducing heritable mutations.
On the other hand, researchers can use an inactivated Cas version that adheres to the target genes and inhibits (CRISPRi) or improves (CRISPRa) the expression of the gene.
The next step in Perturb-seq is to analyze how the perturbation of targeted genes has an impact on the pattern of other kinds of genes expressed by cells. This is achieved by applying a method known as single-cell RNA sequencing (scRNA-seq) that enables a read-out of gene expression from each cell.
In brief, scRNA-seq obtains and labels the gene-expressed molecules (known as messenger RNAs, or mRNAs) from each cell inside a cell population.
With the help of computers, scientists can subsequently read the tags fixed to the mRNA sequences and combine the mRNA identities from every cell. This enables scientists to assess “transcriptomes” or gene expression profiles from every cell to establish how the cells are different from or similar to one another.
Most significantly, in Perturb-seq, the populations of input cells carry CRISPR-based perturbations, and since sgRNAs are integral for mapping the assembled transcriptomes to those CRISPR-based perturbations, it is important to determine the sgRNA identities for every cell.
But standard scRNA-seq methods cannot be used to capture the sgRNAs. Hence, to perform the Perturb-seq experiments, the scientists have earlier depended on indirect mapping techniques but these methods are plagued by technical restrictions. For instance, they are hard to use when multiple sgRNAs are conveyed to every cell.
Adamson and her colleagues, headed by first author Joseph M. Replogle, an MD-PhD trainee at the University of California-San Francisco, wanted to solve these issues.
We developed protocols for capturing sgRNA sequences on different scRNA-seq platforms.”
Britt Adamson, Researcher, Princeton University
These latest procedures, dubbed “direct capture Perturb-seq” by the researchers, offer a method to capture and amplify the sgRNAs along with the cellular transcriptome during scRNA-seq.
Most crucially, direct capture Perturb-seq enables the researchers to easily monitor the presence of numerous sgRNAs in each cell, which the team demonstrated can also prove useful for enhancing CRISPRi.
The authors used the Perturb-seq to duplicate and expand the outcomes of a previous study to demonstrate other methods where direct-capture Perturb-seq could be applied.
The preliminary study, which analyzed the impacts of disrupting pairs of genes on cell growth, showed that blocking expression of specific genes implicated in cholesterol biosynthesis leads to an accumulation of a metabolic intermediate that impairs the cell’s DNA.
Using the direct capture Perturb-seq, the scientists quickly defined how cells react to repression of those genes, to design a model for how cells control the impacts of that intermediate, and to find out what actually happens to cells when they cannot.
As a further enhancement, the researchers also enriched the mRNAs from specific target genes using the scRNA-seq transcriptomes, rendering the scRNA-seq experiments similar to the one explained above, which is cheaper to carry out.
Together, these improvements should help researchers expand the scale of Perturb-seq experiments and better enable efforts to study how genes work.”
Britt Adamson, Researcher, Princeton University
Source:
Journal reference:
Replogle, J. M., et al. (2020) Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nature Biotechnology. doi.org/10.1038/s41587-020-0470-y.