Researchers from Skolkovo Institute of Science and Technology (Skoltech) and their collaborators have studied over 30,000 variants of genetic sequences encoding a pair of fluorescent proteins to determine which traits of mRNA, as well as the characteristics of the first dozen or so codons in it, can enhance the translational efficiency.
An outline of the Escherichia coli experiment. Image Credit: Pavel Odinev/Skolkovo Institute of Science and Technology.
Contrary to some hypotheses, the researchers identified, among other things, that rare codons at the start of the sequence did not appear to improve the translation. The study was published in the Nucleic Acids Research journal.
Translation is one of the underlying processes in any cell, where a ribosome decodes a recently minted messenger RNA (created from DNA during transcription) and forms an amino acid chain that subsequently folds into a protein. This protein, in turn, performs various significant functions in the cell.
Every amino acid is denoted by a codon, which is a triplet of nucleotides in the mRNA chain. There are a total of 61 codons for amino acids but just 20 amino acids can be synthesized by a ribosome, which implies that certain codons are actually synonymous—they encode the same type of amino acid.
Even after many years of research, researchers are still unsure about the level of efficiency at which a cellular “protein plant” works. For example, evidence suggests that some specific secondary structures of mRNA, that is, how the coding sequence gets folded spatially at the beginning, can inhibit the ribosome from attaching to the mRNA and performing its function.
Yet another factor may be associated with those synonymous codons: previous research has indicated the likelihood that codons that are statistically employed more rarely can improve the translational efficiency if positioned at the start of an open reading frame.
Such codons cause the ribosome to move even more slowly along the mRNA at its start so that ribosome queues do not develop downstream.
This is not a futile line of inquiry. Studying the translational efficiency will help one to get a better insight into gene expression and aid biotechnology by ensuring that the protein-producing workhorses of humans are working at their best.
This inspired Ilya Osterman and Zoe Chervontseva of Petr Sergiev, Olga Dontsova and Mikhail Gelfand groups at Skoltech, and Lomonosov MSU and their collaborators to conduct a kind of competition: they examined over 30,000 variants of mRNA encoding the same kind of protein to find out which variant will result in a more efficient translation.
The scientists were interested in the functions of codons number 2 to 11—the first codon is invariably the ATG start codon, more or less like the first line of code in certain programming languages that tells the computer that a program will follow.
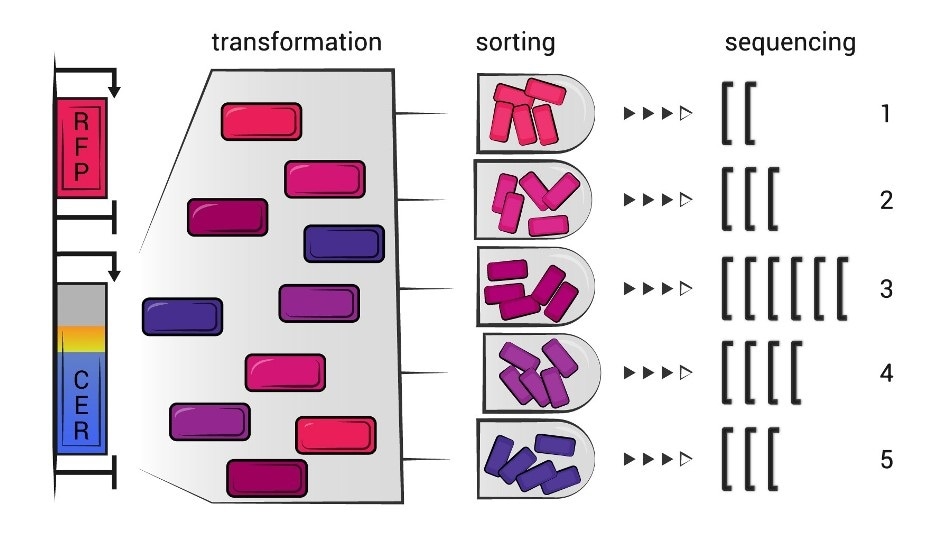
The scientists used Escherichia coli and plasmids—DNA rings encoding the so-called dual fluorescent protein reporter (the “duo” are CER and RFP—two fluorescent proteins). The randomized 30-nucleotide sequences were embedded immediately after the start codon so that they would become codons number 2 to 11 in an mRNA.
Once the modified E. coli was grown to make the RFP and CER proteins and the cells were sorted based on their efficiency, the researchers employed the so-called flowseq approach to establish which coding sequences turned out to be better for efficient production of proteins.
Flowseq is a combination of flow cytometry, a technique where physical and chemical characteristics of cells are measured via the scattering of light from a laser beam, with sequencing of the separated fractions. This method allows to assess the efficiency of protein synthesis in a massively parallel setup, analyzing thousands of variants at a time.”
Ilya Osterman, Principal Research Scientist, Skolkovo Institute of Science and Technology
While the secondary structure of mRNA certainly blocked the translation, the scientists were unable to demonstrate that rare codons at the start of a coding sequence have impacted it in a positive manner.
But they also observed that translation can benefit from additional start codons, whereas the additional Shine-Dalgarno boxes—sequences that help retain the ribosome to the mRNA—are inhibitory.
According to the researchers, their results will help develop more efficient artificial gene constructs that can be used to transform common bacteria, such as E. coli, into potent biotechnological instruments.
Source:
Journal reference:
Osterman, I. A., et al. (2020) Translation at first sight: the influence of leading codons. Nucleic Acids Research. doi.org/10.1093/nar/gkaa430.