Scientists have discovered certain segments of the genetic codes of the SARS and COVID-19 viruses that may support the lifecycle of the viruses. The new methodology is the team’s pioneering tool to find out what genetic sequences preserved as RNA—the chemical cousin of DNA—are actually more stable.
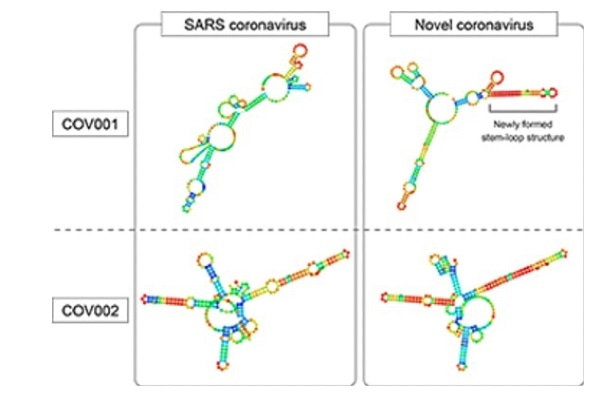
The hairpin shape of these parts of the viruses’ genomes may help the viruses avoid a host’s immune system. Due to a few important mutations, the genome area named COV001 does not form a hairpin structure in the SARS virus (top left), but does in the COVID-19 virus (top right). Image Credit: Image first published in Wakida et al. 2020
The current pandemic is very severe, and we want to contribute to the global acceleration of coronavirus research. Our research includes many types of viruses, but we decided to focus on our coronavirus results.”
Nobuyoshi Akimitsu, Team Lead and Professor, Isotope Science Center, University of Tokyo
The study was carried out by Akimitsu at the University of Tokyo Isotope Science Center. He is an expert in studying how cells are able to tolerate stress.
Several groups of viruses, such as coronaviruses and influenza viruses, preserve their genetic sequence as RNA, which stealthily enters the human cells and deceives them into producing more numbers of viruses. Viruses require their RNA to be stable, withstanding efforts from the immune system of the host to decompose their RNA.
The scientists dubbed their method as Fate-seq because their goal was to find out the fate of a genetic sequence, whether it will survive or degrade depending on its stability.
The Fate-seq technique is a very simple idea. We combined existing technologies in a new way.”
Nobuyoshi Akimitsu, Team Lead and Professor, Isotope Science Center, University of Tokyo
The scientists performed the Fate-seq technique by first splicing a genome into shorter fragments. Even highly dangerous microbes become innocuous when the scientists work only with the shorter, separated, and spliced fragments of their genomes.
The scientists produced RNAs from the fragments of viral genomes and analyzed their fate, or stability, by employing next-generation sequencing. This technique allows the scientists to detect the accurate sequence of individual RNA strands rapidly and concurrently. Computer programs can then be used to detect patterns or fascinating differences in the genetic sequences to explore in more detail.
The scientists examined as many as 11,848 RNA sequences from 26 viral genomes, including the RNA sequence of SARS-CoV. This virus is responsible for causing SARS—the sudden acute respiratory syndrome that killed 774 individuals in the first six months of 2003. The scientists detected a total of 625 stable RNA fragments, of which 21 were from SARS-CoV.
The scientists also made a comparison between the 21 SARS-CoV stable genomic fragments and the full genetic sequence information available for other types of coronaviruses. A couple of the stable fragments from the SARS-CoV were found to be quite common in other evolutionarily analogous coronaviruses, such as the virus responsible for causing COVID-19—SARS-CoV-2.
Predictive models had demonstrated that those two stable fragments of RNA possibly form stem-and-loop structures. Stem-and-loops are short RNA pieces that, rather than remaining in a straight line, tend to fold forward and attach onto themselves, creating a hairpin shape.
In particular, one of the stable RNA fragments forms a stem-and-loop structure only in the COVID-19 virus and not the SARS virus. This is due to the few small yet significant differences in the RNA codes of the viruses.
The stem-and-loop structure of this SARS-CoV-2 genetic fragment is very stable in computer models and we propose that this structure might enhance survival of the virus.”
Nobuyoshi Akimitsu, Team Lead and Professor, Isotope Science Center, University of Tokyo
Apart from gaining a deeper understanding of dangerous viruses, the scientists are hoping to use the Fate-seq technique to figure out the underlying rules of RNA stability and improve novel types of drugs. Human cells employ RNA as an intermediate messenger between protein and DNA.
Engineering RNA-based drugs that are stable and make it easy for cells to translate into protein can potentially treat genetic diseases without the associated dangers of modifying an individual’s DNA.
Source:
Journal reference:
Wakida, H., et al. (2020) Stability of RNA sequences derived from the coronavirus genome in human cells. Biochemical and Biophysical Research Communications. doi.org/10.1016/j.bbrc.2020.05.008.