A protein known as polymerase is used by SARS-CoV-2—the coronavirus that is responsible for causing the worldwide COVID-19 pandemic—to replicate its genome within the infected human cells.
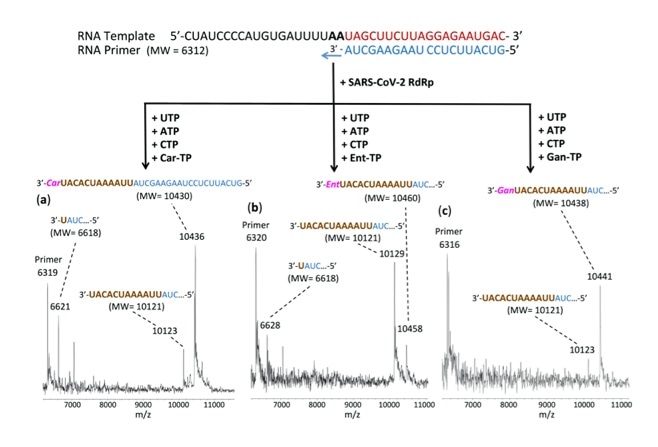
This figure shows that the incorporation of three nucleotide analogues Carbovir-5’-Triphosphate (Car-TP), Entecavir-5’-Triphosphate (Ent-TP), and Ganciclovir-5’-Triphosphate (Gan-TP) by SARS-CoV-2 polymerase terminates the viral polymerase reaction. The reaction products were detected by MALDI-TOF mass spectrometry. Image Credit: Jingyue Ju, Columbia Engineering.
Eliminating the polymerase reaction will halt the growth of this virus, resulting in its eradication by the immune system of the human host.
At Columbia Engineering and the University of Wisconsin-Madison, scientists have discovered an array of molecules that stop the SARS-CoV-2 polymerase reaction, a major step that develops the ability of these molecules as the main lead compounds to be further altered for the development of COVID-19 therapeutics.
Among these, five molecules have already been approved by FDA for use in the treatment of different viral infections, such as hepatitis B, cytomegalovirus, and HIV/AIDS. The new study was published on June 18th, 2020, in the Antiviral Research journal.
The group from Columbia Engineering first reasoned that the active triphosphate of the hepatitis C drug sofosbuvir as well as its derivative could serve as a promising inhibitor of the SARS-CoV-2 polymerase on the basis of the analysis of their molecular properties and also the replication needs of both the coronaviruses and hepatitis C virus.
Under the guidance of Jingyue Ju, Samuel Ruben-Peter G. Viele Professor of Engineering, professor of chemical engineering and pharmacology, and director of the Center for Genome Technology & Biomolecular Engineering at Columbia University, the researchers subsequently teamed up with Robert N. Kirchdoerfer, an assistant professor of biochemistry and also an expert in the analysis of coronavirus polymerases at the University of Wisconsin-Madison’s Institute for Molecular Virology and the department of biochemistry.
In a previous set of experiments in which the properties of the coronavirus polymerase responsible for causing SARS were tested, the scientists observed that the triphosphate of sofosbuvir was capable of eliminating the virus polymerase reaction.
The team subsequently revealed that sofosbuvir as well as four other nucleotide analogs—the active triphosphate forms of the HIV inhibitors Emtricitabine, Tenofovir alafenamide, Alovudine, and Zidovudine—also suppressed the SARS-CoV-2 polymerase with varying levels of efficiency.
Through this molecular insight achieved from these analyses, the researchers developed a method to choose 11 nucleotide analog molecules with a wide range of chemical and structural features as promising inhibitors of the polymerases of both SARS-CoV and SARS-CoV-2.
Although all the 11 molecules tested showed incorporation, six displayed instant termination of the polymerase reaction, three did not eliminate the polymerase reaction, and two displayed delayed termination.
Prodrug medications of five of these nucleotide analogs—Abacavir, Cidofovir, Entecavir, Stavudine, and Valganciclovir/Ganciclovir—that eradicate the polymerase reaction of the SARS-CoV-2 have been approved by the FDA for treating other viral infections. The safety profiles of these medications are also well established.
As soon as the potency of the drugs to suppress viral replication in cell culture is shown in upcoming analyses, the candidate molecules as well as their altered forms could be assessed for developing potential therapies for COVID-19.
In our efforts to help tackle this global emergency, we are very hopeful that the structural and chemical features of the molecules we identified, in correlation with their inhibitory activity to the SARS-CoV-2 polymerase, can be used as a guide to design and synthesize new compounds for the development of COVID-19 therapeutics.”
Jingyue Ju, Department of Chemical Engineering, Center for Genome Technology and Biomolecular Engineering, Columbia University
“We are extremely grateful for the generous research support that enabled us to make rapid progress on this project. I am also grateful for the outstanding contributions made by each member of our collaborative research consortium,” Ju concluded.
Source:
Journal reference:
Jockusch, S., et al. (2020) A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antiviral Research. doi.org/10.1016/j.antiviral.2020.104857.