At the University of Würzburg, researchers are employing mini-organs to model the digestive tract in the laboratory setting. These supposed organoids shed light on the inflammatory processes that lead to diseases like ulcerative colitis and Crohn’s disease.
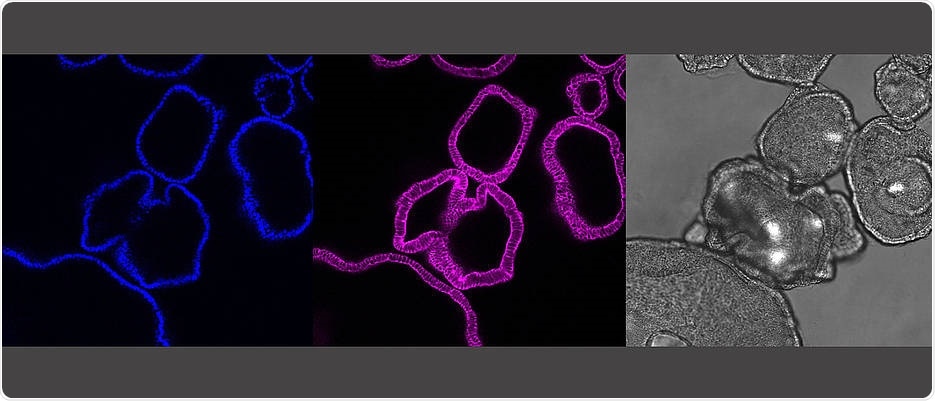
The pictures show the same gut organoids: It shows the cell nuclei (blue) and the skeleton of the cell (pink) as a cross-section of the organoids. In grey is the microscopic picture of the organoids. A single organoid here is about a quarter millimeter in size. Image Credit: Sina Bartfeld/Universität Würzburg.
Researchers do not know why certain chronic inflammatory bowel diseases, like Crohn’s disease, have an impact on both the colon and the small intestine, whereas others, like ulcerative colitis, are limited to the colon.
To solve clinical mysteries like this one, among others, scientists from the University of Würzburg have developed tiny models of the digestive tract in the laboratory. One of the researchers’ findings is that the digestive tract includes an intrinsic segmentation that could provide insights into these common inflammatory diseases.
At present, researchers are able to create tiny versions of virtually any organ of human bodies—including the intestine, brain, and skin. Such three-dimensional (3D) structures are produced from stem cells known as “organoids.”
Organoids have a diameter of about 0.5 mm, perhaps resembling the size of a mustard grain; however, organoids have a striking similarity to the actual organs.
Despite their miniscule size, organoids simulate the organ they are derived from extremely well. Organoids contain the same types of cells as the real organ. The stem cells from which the organoids are generated contain a kind of pre-programmed tissue identity. The stem cell ‘knows’ which organ it comes from and even in culture it produces the kinds of cells that are present in this organ in our bodies.”
Dr Sina Bartfeld, Study Lead, Research Center for Infectious Diseases, Institute for Molecular Infection Biology
Dr Bartfeld’s research team, in association with surgeon Armin Wiegering from the University Hospital of Würzburg, produced organoids from the colon, small intestine, and stomach. To their amazement, the researchers found a huge molecular complexity, as shown by RNA sequencing, which mirrors the cells’ gene activity.
One of the researchers’ results was that organoids from the varying segments of the digestive tract turn on particular gene-programs, based on their tissue identity.
It’s intuitive that gastric and intestinal cells have to produce different digestive enzyme—but we were surprised to discover that particular binding sites of the immune system are also part of this tissue identity.”
Dr Sina Bartfeld, Study Lead, Research Center for Infectious Diseases, Institute for Molecular Infection Biology
The specific assembly of the immune binding locations may have a role to play in the inflammatory diseases specific to organs. It may even be applicable to cancer development, in which chronic inflammation has also been implicated.
But whether this is the case and how inflammation is likely to play a role in carcinogenesis needs more research, for which organoids form a new groundwork.
Organoids can be produced quickly and in large numbers in the laboratory. They also have the added benefit of containing human tissues and providing a rough representation of a human organ.
Since considerable variations exist between animals and humans, organoids can help decrease animal experiments and shed light on uniquely human disorders. Organoids also play an increasingly significant role in the development of drugs.
Organoids demonstrate the amazing organization of the gut—also regarding the recognition of viruses and bacteria
Moreover, organoids provide a completely new way of examining the underlying molecular procedures that occur in a biologically realistic model—like the digestive system, which is also the target of Dr. Bartfeld’s research group at the University of Würzburg.
The epithelial cells that line the human digestive tract play a major barrier function, which inhibits microbes from entering the human bodies. These may be pathogens, like disease-causing viruses or bacteria.
The intestine is colonized simultaneously by an unlimited number of beneficial bacteria—the supposed microbiota, which helps human to digest food. Hence, the epithelial cells should be able to perceive both hostile and friendly viruses or bacteria and respond suitably. This is achieved via unique binding sites, known as pattern recognition receptors.
Such receptors identify particular molecules created by the different microorganisms in the intestine. If the epithelial cells identify molecules created by harmful microorganisms, as opposed to the beneficial bacteria, they have to trigger an alarm and promote an immune response. To date, it is not completely known how the epithelium is able to distinguish between foe and friend.
According to Dr. Bartfeld, “It is extremely difficult to untangle the complex interaction between immune cells, epithelial cells, and microbes. However, since our organoids contain only epithelial cells, we can use them to specifically investigate the contribution of the epithelium in this interaction.”
During their research, the researchers found out that every pattern recognition receptor has its individual, segment-specific gene activity pattern.
The stomach as well as each segment of the intestine has its own specific repertoire of pattern recognition receptors. Thus, the immune response of the epithelium is location-specific. In this way, the stomach reacts to different bacterial compounds than the small intestine or the colon.”
Özge Kayisoglu, Study First Author, University of Würzburg
Such variances in the immune response may play a role in segment-specific disorders like Crohn’s disease or ulcerative colitis.
It was not known what exactly triggers this differential reaction in bacterial compounds. Originally, the evident assumption was that the immune receptors are controlled in response to colonization with beneficial bacteria. The researchers tested this theory by creating organoids that had never come into contact with microorganisms.
“The data showed that the microbiota does have an effect—but we were surprised to find that in large part, immune recognition of the epithelium is in fact genetically determined during development and independent of the environment,” Bartfeld concluded.
On the whole, the researchers’ findings represent a significant step in shedding light on inflammatory processes. These findings reveal that every part of the digestive tract has its individual specific combination of immune recognition receptors. Such innate immunity dysfunctions may encourage the development of inflammatory disorders.
Source:
Journal reference:
Kayisoglu, O., et al. (2020) Location-specific cell identity rather than exposure to GI microbiota defines many innate immune signaling cascades in the gut epithelium. Gut. doi.org/10.1136/gutjnl-2019-319919.