Proteins regulate several physiological functions that are crucial for living things in vivo. More than 20,000 protein types are found in human beings, and every protein has a different role to play by interacting with other types of proteins.
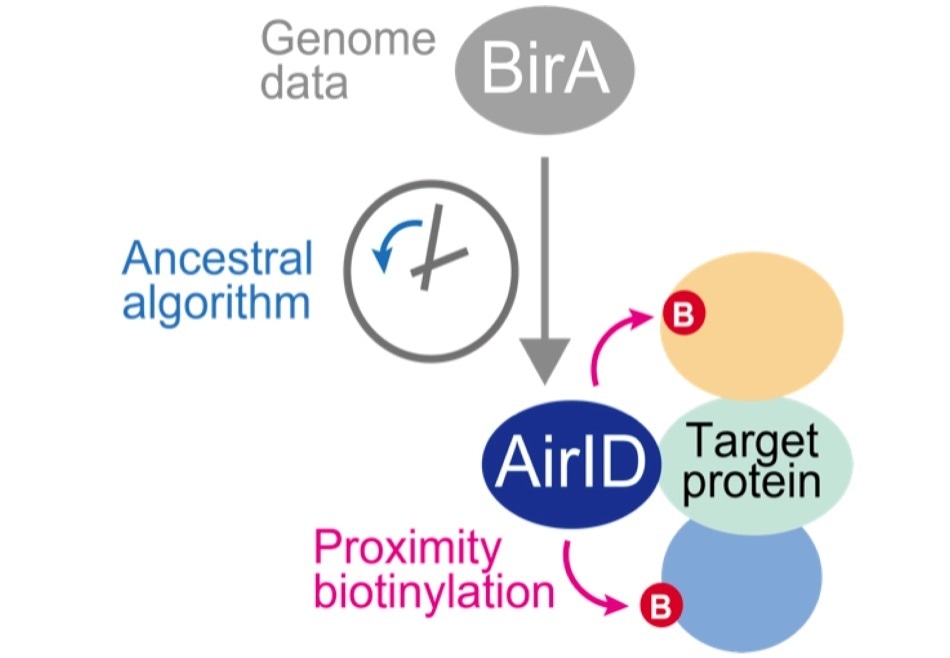
An ancestral type enzyme AirID was designed from the sequence of biotinylation enzyme BirA of Escherichia coli by an ancestral enzyme reconstruction algorithm with metagenome data. Since AirID has high biotinylation activity and low cytotoxicity, it is possible to biotinylate a protein that interacts with the target protein by expressing it in cells as a fusion protein with the target protein. By identifying these biotinylated proteins using mass spectrometry analysis, it becomes possible to comprehensively identify proteins that interact with the target protein in cells. Image Credit: Tatsuya Sawasaki (Ehime University).
Proximity-dependent biotin identification (BioID) technology is a method used for detecting such interacting partner proteins. In the BioID technique, the BioID-fusion protein should be expressed and biotin should be added.
In cells that express BioID-fusion bait protein, proteins with which the BioID-fusion bait protein communicate are biotinylated and can be thoroughly examined through precipitation with streptavidin and then by mass spectrometry.
To date, three types of BioID enzymes, namely TurboID, BioID, and BioID2, have been created. However, TurboID also biotinylates the non-interacting proteins, while both BioID2 and BioID need a long reaction time. Additional enhancements of biotinylation enzymes are a significant objective for improving the ease of proximity biotinylation in cells.
In this study, the researchers used metagenome data and an ancestral enzyme reconstruction algorithm to create ancestral biotinylation enzymes. The novel enzyme—ancestral BirA for proximity-dependent biotin identification, or AirID for short—has demonstrated higher specificity, higher activity, and less toxicity when compared to the three previous types of enzymes.
AirID was used for assessing protein interaction inhibitors, examining complexes developed by three or more protein types, and examining interactions through compounds known as Molecular Glue.
Additionally, the scientists have successfully and thoroughly identified proteins that communicate with a target protein by detecting the biotinylated proteins through mass spectrometry analysis in cultured cells. These cells express the target protein merged with the AirID enzyme.
These findings show that the AirID is an appropriate enzyme for examining detailed protein–protein interactions that take place in cells.
Source:
Journal reference:
Kido, K., et al. (2020) AirID, a novel proximity biotinylation enzyme, for analysis of protein–protein interactions. eLife. doi.org/10.7554/eLife.54983.