Researchers from the Korea Advanced Institute of Science and Technology (KAIST) have employed an X-ray technique to monitor the mechanism of protein folding. This could enhance the computer simulations of this process and hold implications for enhancing drug discovery and interpreting various diseases.
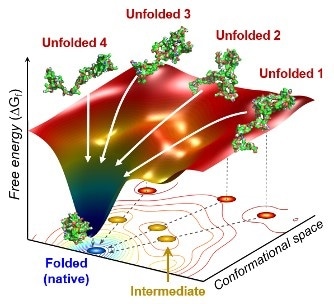
The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state. Image Credit: Professor Hyotcherl Ihee, The Korea Advanced Institute of Science and Technology.
The study results were reported in the Proceedings of the National Academy of Sciences of the United States of America on June 30th, 2020.
Whenever proteins are converted from their DNA codes, they rapidly change from an unfolded, non-functional state into their folded, functional state. Folding problems can result in various diseases, such as Parkinson’s and Alzheimer’s.
Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure.”
Hyotcherl Ihee, Physical Chemist, Department of Chemistry, Korea Advanced Institute of Science and Technology
Dr Tae Wu Kim, the study’s lead author from Ihee’s team, stated, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Using an X-ray scattering method, Ihee’s research team revealed the folding of the protein—cytochrome c—from its initial unfolded state. Such a protein includes a chain of 104 amino acids along with an iron-containing heme molecule. This protein is mostly employed in protein folding research.
After placing this protein in a solution, the scientists shined ultraviolet light on it. This procedure supplies electrons to the cytochrome c protein, reducing the iron inside it from the ferric to the ferrous state, which triggers the folding process.
While this was occurring, the researchers radiated X-rays at extremely short intervals onto the specimen. Following this, the X-rays scattered off all the pairs of atoms in the specimen and a detector constantly captured the scattering patterns of the X-rays.
The X-ray scattering patterns offered direct data about the three-dimensional (3D) structure of proteins, and the modifications made in these patterns over time revealed the real-time movement of the protein at the time of the folding process.
The researchers observed that cytochrome c proteins first exist in a wide range of unfolded states. After the folding process is activated, these proteins stop by a cluster of intermediates within 31.6 µs and those intermediates subsequently follow different kinds of routes with different folding times to achieve a folded state that is energetically stable.
We don’t know if this diversity in folding paths can be generalized to other proteins. However, we believe that our approach can be used to study other protein folding systems.”
Hyotcherl Ihee, Physical Chemist, Department of Chemistry, KAIST University
Ihee believes that this method can enhance the precision of models that mimic the interactions of protein by including data on their unstructured states. Such simulations are significant because they can help detect obstacles to proper folding and estimate the folded state of a protein based on its sequence of amino acids. Eventually, the models could help explain the development of certain diseases and the interaction of drugs with numerous protein structures.
Source:
Journal reference:
Kim, T. W., et al. (2020) Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering. Proceedings of the National Academy of Sciences. doi.org/10.1073/pnas.1913442117.