DNA is considered the storage unit of all genetic data in living organisms. This data is used to encode proteins, subsequently allowing biological systems to work as required for the organism to live.
Tailored DNA microstructures. Changing the DNA sequence of the sticky ends in the Y-motifs allows for obtaining DNA droplets with various useful and highly designable behaviors. Image Credit: Tokyo Institute of Technology.
The functioning of the DNA is facilitated by its structure: a double-stranded helix formed through the merging of certain pairs of molecules known as “nucleotides” in specific orders, referred to as “sequences.”
In recent years, researchers in the domains of DNA nanotechnology were able to engineer DNA sequences to build the required microstructures and nanostructures, which can be used for producing artificial cell systems or for analyzing biomolecular functions.
Thanks to the customization of the designs of sequences in DNA nanotechnology, the communications among DNA molecules can be regulated and programmed. The inter-molecular communications in cells result in numerous phenomena.
A phenomenon known as “liquid-liquid phase separation (LLPS)”—the isolation of a liquid into a denser phase of droplets inside a more dilute phase—has a significant role to play in many biological processes. LLPS artificially produced through DNA nanotechnology can provide a better insight into the applicability of LLPS and offer a framework for regulating bio-macromolecular droplets.
Thus, a research team from the Tokyo Institute of Technology, headed by Professor Masahiro Takinoue, developed certain DNA nanostructures to figure out the effect of DNA sequences and show controllability on LLPS—into DNA-poor and DNA-rich phases—in artificially developed DNA nanostructures.
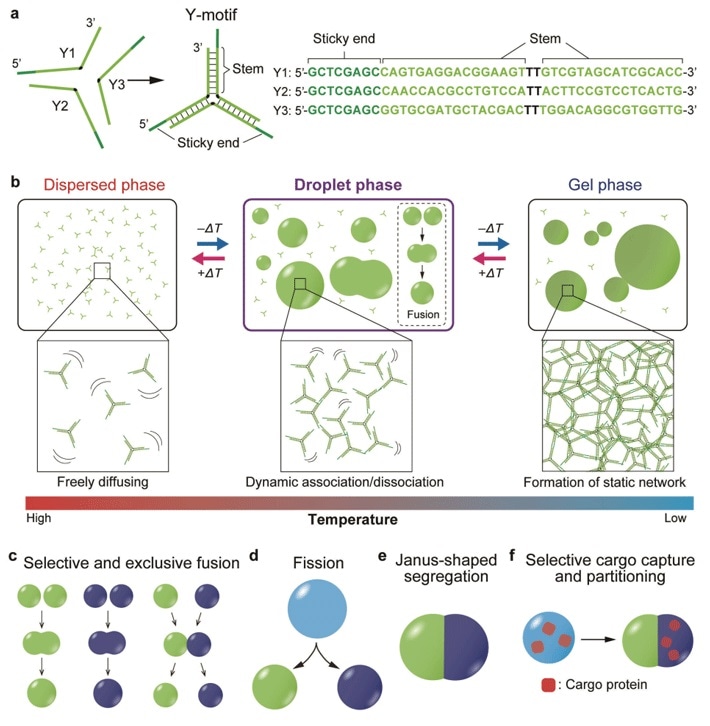
Published in the Science Advances journal, the study involved the development of Y-shaped DNA nanostructures known as “Y-motifs.”
Every side of a Y-motif contains a short sticky end that communicates with other “compatible” sticky ends. By gradually reducing the temperature, the researchers discovered that the Y-motifs reversibly agglomerate to create droplets and later gels.
When the researchers added another set of the constructed Y-motifs with sticky ends that could not be used with the earlier set, it resulted in the formation of two sets of droplets for every type of Y-motif. This showed that sequences of DNA can be customized to combine entirely with analogous ones.
Subsequently, Professor Takinuoe and collaborators designed a unique DNA structure that can bridge the incompatible Y-motifs together. When this was added to the Y-motif mixture, droplets made up of both motifs were formed.
Additional development of a cleavable variant of the exclusive bridge DNA structure and subsequent addition of a specific cleaving enzyme caused the mixed droplets and the fission of droplets to separate into Janus-shaped droplets with immiscible halves comprising both types of Y-motif.
When the researchers conjugated the cargo molecules with DNA strands that were compatible with either type of Y-motif, they successfully localized the cargo molecules solely on one half of the droplet.
The researchers were thus able to “program” the DNA and “control” their behavior, paving the way for a novel method for producing artificial reaction settings to analyze drug delivery and biological systems.
Living systems are well-organized dynamic structures whose behavior is regulated by the information encoded in biopolymers (such as DNA). Our DNA-based liquid-liquid phase separation system could provide a new basis for the development of artificial cell engineering.”
Masahiro Takinoue, Study Corresponding Author and Professor, Department of Computer Science, Tokyo Institute of Technology
Since the exact DNA sequences can be easily created using the existing bioengineering methods, the promising applications of exploiting material behaviors via DNA sequences are far-reaching.
The phase behavior shown in this study could be expanded to other materials that can be modified with DNA, which may enable us to design phases and create droplets for various materials. Moreover, we envision that the observed autonomous behavior of macromolecular structures could one day serve for the development of robotic molecular systems comparable to those of living cells.”
Masahiro Takinoue, Study Corresponding Author and Professor, Department of Computer Science, Tokyo Institute of Technology
Source:
Journal reference:
Sato, Y., et al. (2020) Sequence-based engineering of dynamic functions of micrometer-sized DNA droplets. Science Advances. doi.org/10.1126/sciadv.aba3471.