The lives of millions or even billions of individuals across the world have been changed by the ongoing COVID-19 pandemic. COVID-19 is a disease caused by an RNA virus called SARS coronavirus 2 (SARS-CoV-2), that is, a virus that employs RNA to preserve its genetic data.
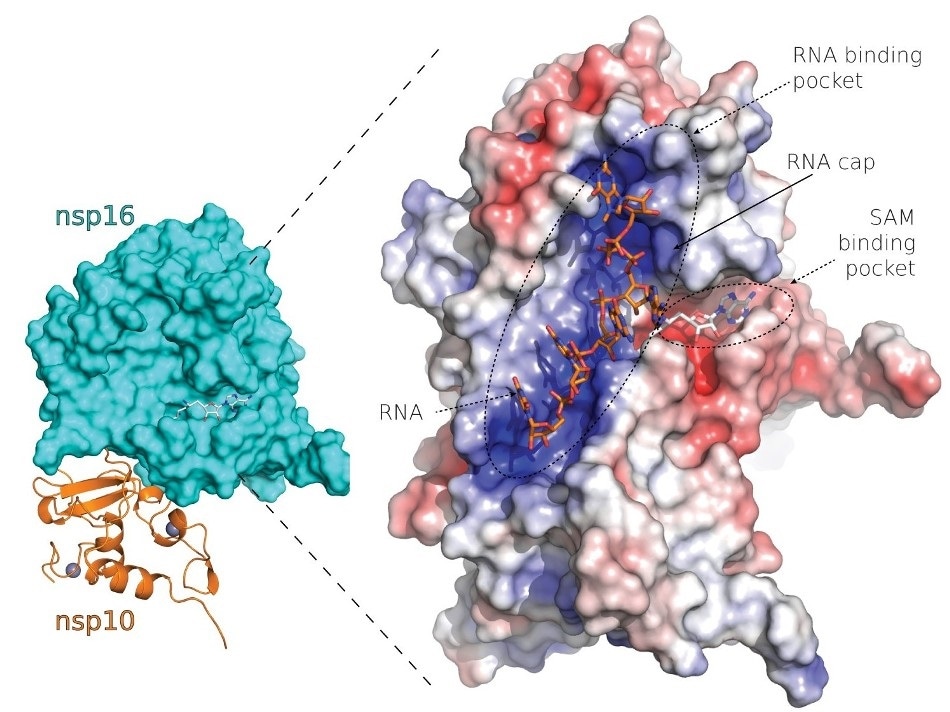
SARS-CoV-2 nsp10-nsp16 protein complex. Image Credit: Petra Krafcikova/Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences.
To tackle this virus, one should have a thorough understanding of the function and structure of its individual proteins.
The coronavirus uses several mechanisms to try and outwit the human immunity and convinces the human cells that the viral RNA is actually innocuous.
One such mechanism used by the virus is to install the so-called unique structures at the start of the RNA, due to which the viral RNA mimics the human RNA and enables the virus to infect the human body and grow in numbers inside it.
The Nsp16 is a coronavirus protein that is catalyzed by the cap installation process, where another viral protein called Nsp10 is also involved.
A research team, from the Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences and headed by Dr Evžen Bouřa and Dr Radim Nencka, employed X-ray crystallography to establish and study the accurate structure of the complex of both these proteins.
This analysis enabled scientists to detect many underlying properties of the Nsp10 and Nsp16 protein complex, such as a deep canyon present on the surface of the protein complex. The viral RNA binds at this site of deep canyon, followed by cap installation.
Inhibitors that block the activity of the Nsp10 and Nsp16 protein complex and thus suppress the activity of the whole cap installation process can target this deep canyon. This canyon could be used as drugs to fight several coronaviruses in the days to come.
Hundreds of research teams tried to shed light on how the COVID-19 virus is able to hide its RNA from cellular immunity. In the end, two American teams and we here in Prague were the ones who succeeded. We used X-ray analysis to determine the structure of the responsible viral enzyme with inhibitor.”
Evžen Bouřa, Head of the Structural Membrane Biology Group, Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences
The team has published the study results in Nature Communications, a leading journal.
Source:
Journal reference:
Krafcikova, P., et al. (2020) Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nature Communications. doi.org/10.1038/s41467-020-17495-9.