Legume plants can recognize their friends from their foes, and now scientists have found out how they do it at the molecular level.
Scientists have discovered that legumes use small, well-defined motifs in LysM receptors to read signals produced by both pathogenic and symbiotic microbes. These findings in Science have enabled the researchers to reprogram the chitin immune receptor into a symbiotic receptor. Image Credit: Christina Krönauer and Damiano Lironi.
Plants detect beneficial microorganisms and keep out the harmful ones, which is significant for the healthy production of plants and thus for global food security.
Now, researchers have found how legumes utilize tiny and well-defined motifs present in receptor proteins to read molecular signals created by both symbiotic and pathogenic microorganisms.
Such incredible discoveries have allowed the team to reprogram immune receptors into symbiotic receptors, representing the first milestone for designing symbiotic nitrogen-fixing symbiosis into cereal crops.
Legume plants use the help of symbiotic bacteria, known as Rhizobia, to fix the nitrogen in the air. These bacteria colonize the roots of these legume plants. Hence, plants should have the ability to accurately detect their symbiont to prevent infection caused by pathogenic microorganisms.
As such, legumes employ different types of LysM receptor proteins that are situated on the external cell surface of their roots.
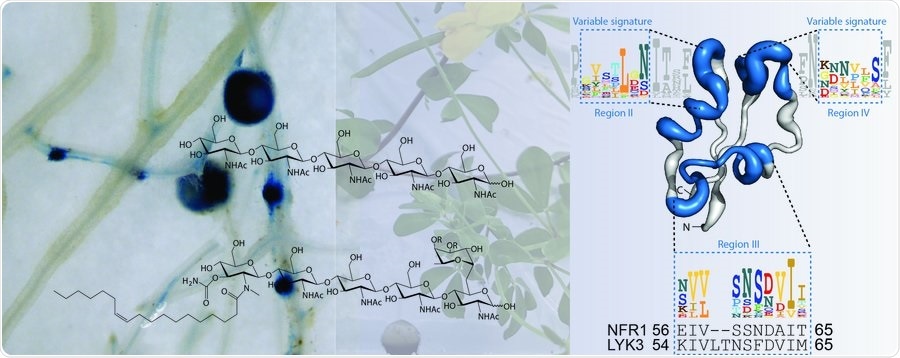
Headed by Aarhus University, an international research team demonstrated that symbiotic or (Nod factors) or pathogenic (chitin) signaling molecules are detected by tiny molecular motifs on the receptors that guide the signaling output towards antimicrobial defense or symbiosis. The study was published in the Science journal.
LysM receptors are found in all land plants. They ensure the detection of numerous microbial signals, but scientists are not clear how a plant decides to mount an immune response or a symbiotic towards an incoming microorganism.
We started by asking a basic and, maybe at start, naïve question: Can we identify the important elements by using very similar receptors, but with opposing function as background for a systematic analysis?”
Zoltán Bozsoki, Department of Molecular Biology and Genetics, Aarhus University
Kira Gysel added, “The first crystal structure of a Nod factor receptor was a breakthrough. It gave us a better understanding of these receptors and guided our efforts to engineer them in plants.”
The research integrates the structure-assisted dissection of distinct regions in planta functional analysis and in LysM receptors for biochemical experiments.
To really understand these receptors, we needed to work closely together and combine structural biology and biochemistry with the systematic functional tests in plants.”
Simon Boje Hansen, Department of Molecular Biology and Genetics, Aarhus University
The scientists used this method to detect previously unfamiliar motifs in the LysM1 domain of Nod factor and chitin receptors as determinants for symbiosis and immunity.
It turns out that there are only very few, but important, residues that separate an immune from a symbiotic receptor and we now identified these and demonstrate for the first time that it is possible to reprogram LysM receptors by changing these residues.”
Kasper Røjkjær Andersen, Department of Molecular Biology and Genetics, Aarhus University
The long-term objective is to shift the special nitrogen-fixing potential of the legume plants into cereal plants to restrict the need to use polluting commercial nitrogen fertilizers and to empower and benefit the poorest people living on Earth.
“We now provide the conceptual understanding required for a stepwise and rational engineering of LysM receptors, which is an essential first step towards this ambitious goal,” Simona Radutoiu concluded.
Source:
Journal reference:
Bozsoki, Z., et al. (2020) Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science. doi/10.1126/science.abb3377