Genome sequencing of plants, bacteria, and even human beings has turned out to be a standard process, but despite this fact, the genome continues to raise several unanswered questions.
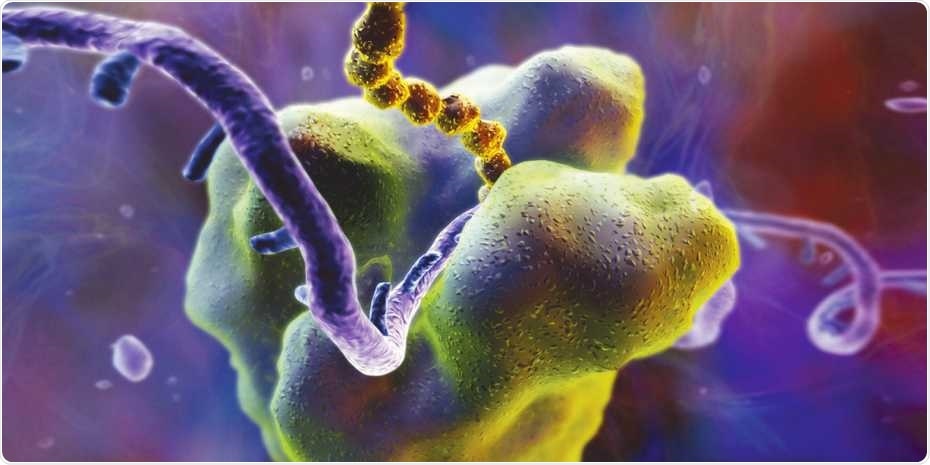
Ribosomes are the cell’s protein-making machinery. They read the genetic information encoded in messenger RNA (violet) and translate it into proteins (yellow). Image Credit: Science Photo Library.
One of these questions is related to the locations on messenger RNAs, or mRNAs, that ribosomes attach to translate the genetic data, where ribosomes are cellular structures involved in protein synthesis. At present, the role of these ribosome binding locations has been only partially understood.
Now, an interdisciplinary research team from the Department of Biosystems Science and Engineering (D-BSSE) at ETH Zurich in Basel has designed a novel method that helps acquire in-depth data on a remarkably huge number of these binding locations in bacteria, for the first time. The novel method integrates machine learning with experimental methods of synthetic biology.
Precise control over protein production
The binding sites of ribosomes are essentially short RNA sequences upstream of a gene’s coding sequence. In earlier days, biotechnologists also designed synthetic binding locations. While the ribosomes attach extremely well to some of these sites, they do not attach quite well to other locations.
The tighter ribosomes are capable of attaching to a particular variant—that is, if the ribosomes increasingly translate the respective gene, they tend to produce more amount of the corresponding protein.
Biotechnologists who make use of bacteria to synthesize target chemicals, like pharmaceuticals, can control the quantity of proteins involved in the cell by choosing their ribosome binding locations.
Exerting this kind of control is particularly important and helpful when incorporating complex gene networks comprising multiple proteins at the same time. The key here is to establish an optimal balance amongst the different proteins.”
Markus Jeschek, Senior Scientist and Group Leader, ETH Zurich
An experiment with 300,000 sequences
Along with ETH professors Yaakov Benenson and Karsten Borgwardt and also with members of the respective teams, Jeschek has currently developed a new approach to establish the level of tightness at which the ribosomes attach to hundreds of thousands or more sequences of RNA in a single experiment. Earlier, this could be achieved only for a few hundred sequences.
The method developed by ETH scientists harnesses deep sequencing—the cutting-edge technology used for sequencing both RNA and DNA. In a laboratory setting, the researchers created more than 300,000 different synthetic ribosomal binding locations and combined each of these with a gene for an enzyme that alters a part of the target DNA.
The team introduced the resulting gene constructs into bacteria to observe how tightly can the ribosomes attach to RNA in each separate case. Improved function of the binding location resulted in increased production of the enzyme in the cell and also caused the target DNA to change more quickly.
Towards the end of the experiment, this modification can be read along with the binding site of the RNA sequence through deep sequencing.
Universally applicable approach
As 300,000 different synthetic ribosome binding sites denote only a small part of the several billions of theoretically viable ribosome binding locations, the researchers used machine learning algorithms to examine their data.
These algorithms can detect complex patterns in large datasets. With their help, we can predict how tightly ribosomes will bind to a specific RNA sequence.”
Karsten Borgwardt, Professor of Data Mining, ETH Zurich
Thanks to the ETH scientists, this prediction model is freely available as software so that it can be used by other investigators as well. The team will shortly introduce a user-friendly online service.
The researchers’ method is universally relevant, emphasized Benenson and Jeschek, and the research team has planned to extend it to other organisms, such as human cells.
We’re also keen to find out how genetic information influences the amount of protein that is produced in a human cell. This could be particularly useful for genetic diseases.”
Yaakov Benenson, Professor, ETH Zurich
Source:
Journal reference:
Höllerer, S., et al. (2020) Large-scale DNA-based phenotypic recording and deep learning enable highly accurate sequence-function mapping. Nature Communications. doi.org/10.1038/s41467-020-17222-4.