Vertebrate organs are known to manage physiological activities, and the varied expression patterns of scores of genes govern the functions and identities of organs.
Complex evolutionary relationships: Long-term expression in one organ predisposes genes for later use in other organs. Image Credit: Kenji Fukushima/University of Würzburg.
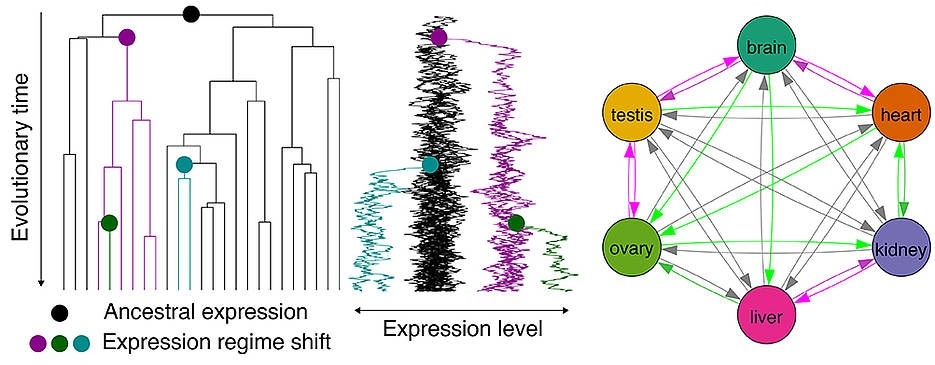
Due to this mechanism, the evolution of gene expression patterns has a primary role to play in organismal evolution.
The development of the hominoid brain, which is one of the crucial organ-altering evolutionary events, is also linked to the shifts in gene expression. While gene duplication is known to play a significant role in expression pattern shifts, the evolutionary dynamics of expression patterns both with and without the duplication of genes is yet to be clearly understood.
An important question is whether long-term expression in one organ predisposes genes to be subsequently utilized in other organs. The answer is yes, there are preadaptive propensities in the evolution of vertebrate gene expression, and the propensity varies with the presence and type of gene duplication.”
Dr Kenji Fukushima, Julius-Maximilians University of Würzburg
Funded by the Alexander von Humboldt Foundation
Now, Kenji Fukushima has reported this and other discoveries along with the study’s co-author David D. Pollock from the University of Colorado, School of Medicine, Aurora, United States, in the Nature Communications journal.
Dr Fukushima maintains a research position at the Chair for Molecular Plant Physiology and Biophysics of the Julius-Maximilians University of Würzburg. Since 2018, this Japanese evolution biologist has been setting up a working team, financially supported with 1.6 million euros by the Alexander von Humboldt Foundation.
The eminent foundation had chosen Dr Fukushima as the winner of its Sofja Kovalevskaja Prize 2018. The award is meant for remarkably talented young researchers.
Complex history of gene family trees
The researchers amalgamated a total of 1,903 RNA-seq datasets from 182 research projects for their study. The datasets include six organs (the heart, brain, liver, kidneys, testis, and ovary) from 21 vertebrate species, which include birds, lizards, freshwater fish and frogs, humans, and rodents. Therefore, the team was able to unravel a complex history of gene family trees. This also enabled them to study the evolutionary expression of a wide set of genes.
Source:
Journal reference:
Fukushima, K & Pollock, D D (2020) Amalgamated cross-species transcriptomes reveal organ-specific propensity in gene expression evolution. Nature Communications. doi.org/10.1038/s41467-020-18090-8.