Cancer driver genes have modifications that are crucial for the development and proliferation of tumor.
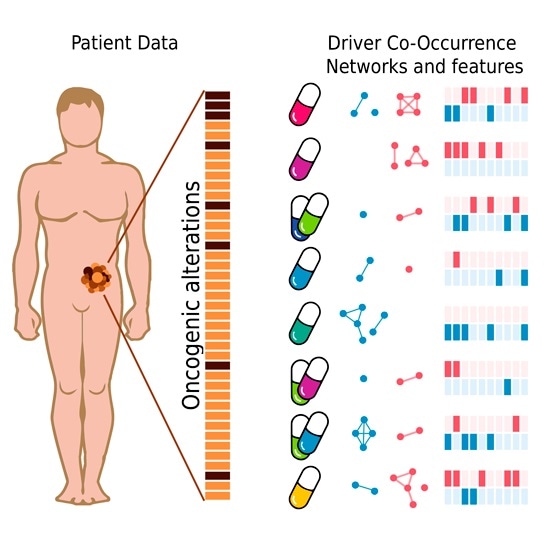
A collection of drug response classifiers based on DCO networks. Image Credit: SBNB lab (IRB Barcelona).
Scientists from the Structural Bioinformatics and Network Biology (SBNB) Laboratory at IRB Barcelona, and headed by ICREA researcher Patrick Aloy, have now devised a computational pipeline that estimates the response of tumor to different cancer therapies.
The new system is built on the detection of complex response markers obtained from the patterns of co-occurrence between cancer driver genes that carry mutations. The system has been tested at the experimental level and, with information collected from breast cancer patients, has achieved an accuracy of 66% in the prediction of such responses.
Cancer driver genes, which play a vital role in the development of tumors, have been extensively researched in the last several years. A better understanding of which of these genes are likely to be affected in a certain tumor can help spot the most suitable therapeutic strategy for that patient, through a method called precision medicine.
For the first time, the SBNB Lab team has proposed the co-occurrence (or lack thereof) of modifications in two or more cancer driver genes as a crucial factor in estimating the response to a particular therapy.
The sum of two or more cancer driver genes affected by mutations leads to the formation of a complex network of biomarkers, alters the molecular profile of the tumor and affects its response to treatments. Through this work, we see that studying cancer driver genes as a whole, analyzing the different combinations, can bring about a great advance towards precision medicine.”
Patrick Aloy, Researcher, ICREA
From bioinformatics to experimental and clinical analysis
While there is plenty of information on cancer genomes, not much data is available on the result of therapeutic interventions in patients.
The team began from a public database that gathers data on the effect of numerous therapies on the development of human tumors that have been implanted in a mouse model. On the basis of this information, the team chose 53 treatments or combinations of therapies and compared the molecular profiles of tumors that reacted to every treatment and those that did not.
After developing our computational model, we validated it experimentally in human tumours implanted in mice. We were able to predict the outcome of the therapy in 12 of the 14 case studies, well above the power of approved biomarkers to predict drug response.”
Lídia Mateo, Study First Author and Postdoctoral Researcher, SBNB Laboratory, IRB Barcelona
The team also confirmed the algorithm with the treatment response information obtained from breast cancer patients.
Source:
Journal reference:
Mateo, L., et al. (2020) Personalized cancer therapy prioritization based on driver alteration co-occurrence patterns. Genome Medicine. doi.org/10.1186/s13073-020-00774-x.