For the first time, scientists have sequenced the genome of Alexander Fleming’s penicillin mould and compared it to the later models.
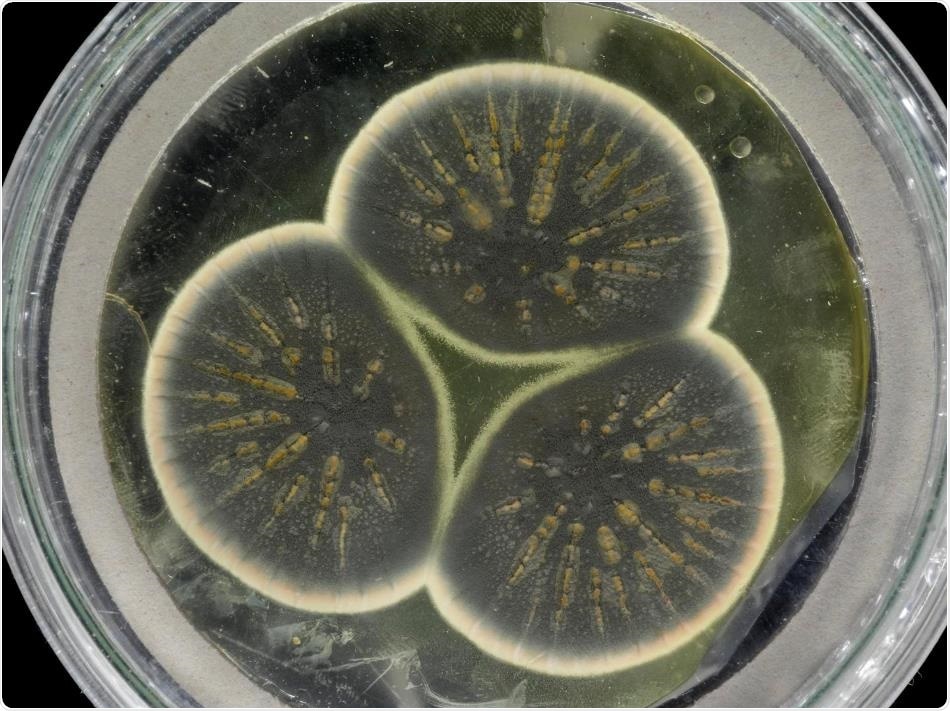
Mould regrown from Fleming's frozen sample. Image Credit: CABI.
The first antibiotic—penicillin—was discovered by Alexander Fleming in 1928 while he was working at St Mary’s Hospital Medical School, which is presently a part of Imperial College London. The antibiotic was synthesized by a mould in the genus Penicillium that inadvertently began to grow in a Petri dish.
The team from Imperial College London, CABI, and the University of Oxford have now sequenced the genome of Fleming’s original strain of Penicillium using specimens that were frozen alive over 50 years ago.
The researchers also employed the new genome to make a comparison of Fleming’s mould with two Penicillium strains from the United States that are used to synthesize the antibiotic on a large scale.
The study results, recently published in the Scientific Reports journal, demonstrate that the US and UK strains of Penicillium use slightly different techniques to synthesize penicillin, possibly indicating new routes for large-scale production.
Historical significance
We originally set out to use Alexander Fleming’s fungus for some different experiments, but we realised, to our surprise, that no-one had sequenced the genome of this original Penicillium, despite its historical significance to the field.”
Timothy Barraclough, Lead Researcher and Professor, Department of Life Sciences, Imperial College London
Barraclough also worked in the Department of Zoology at Oxford.
While Fleming’s mould is known to be the original source of penicillin, large-scale production rapidly moved to using fungus from mouldy cantaloupes in the United States. And from these natural beginnings, the samples of Penicillium were artificially chosen for strains that create higher amounts of penicillin.
The researchers re-cultured Fleming’s original Penicillium from a frozen specimen preserved at the culture collection at CABI and isolated the DNA for sequencing. They later compared the resulting genome with the earlier published genomes of a pair of industrial Penicillium strains used later in the United States.
The team specifically observed two kinds of genes: those that control the enzymes, for instance, by regulating the number of enzymes that are produced; and those encoding the enzymes that are used by the fungus to synthesize penicillin.
The regulatory genes in both the US and UK strains had the same genetic code; however, there were more copies of the regulatory genes in the US strains, assisting those strains to synthesize more quantities of penicillin.
But the genes coding for penicillin- producing enzymes varied between the strains separated in the US and UK. According to the team, this demonstrates that the evolution of wild Penicillium in the UK and US occurred naturally to create slightly varied versions of these enzymes.
Microbial arms race
Penicillium is one of the molds that create antibiotics to combat bacteria and is in a constant arms race since microbes evolve new ways to escape from these defenses. The UK and US strain probably evolved differently to adapt to their local microorganisms.
The evolution of microbes is a big issue today, with many becoming resistant to antibiotics. While the researchers stated that they are yet to know the outcomes of the sequences of different enzymes in the US and UK strains for the eventual antibiotic, it certainly raises the intriguing prospect of identifying new ways to alter the production of penicillin.
Our research could help inspire novel solutions to combatting antibiotic resistance. Industrial production of penicillin concentrated on the amount produced, and the steps used to artificially improve production led to changes in numbers of genes.”
Ayush Pathak, Study First Author, Department of Life Sciences, Imperial College London
“But it is possible that industrial methods might have missed some solutions for optimizing penicillin design, and we can learn from natural responses to the evolution of antibiotic resistance,” Pathak concluded.
Source:
Journal reference:
Pathak, A., et al. (2020) Comparative genomics of Alexander Fleming’s original Penicillium isolate (IMI 15378) reveals sequence divergence of penicillin synthesis genes. Scientific Reports. doi.org/10.1038/s41598-020-72584-5.