There is currently an urgent need to develop fast, reliable, and scalable detectors for the screening of SARS-CoV-2. A recent paper published on the medRxiv* preprint server (October 2020) by Santiago-Frangos et al. describes a repurposed type III CRISPR-Cas system that can detect the virus from nasopharyngeal swabs in less than an hour.
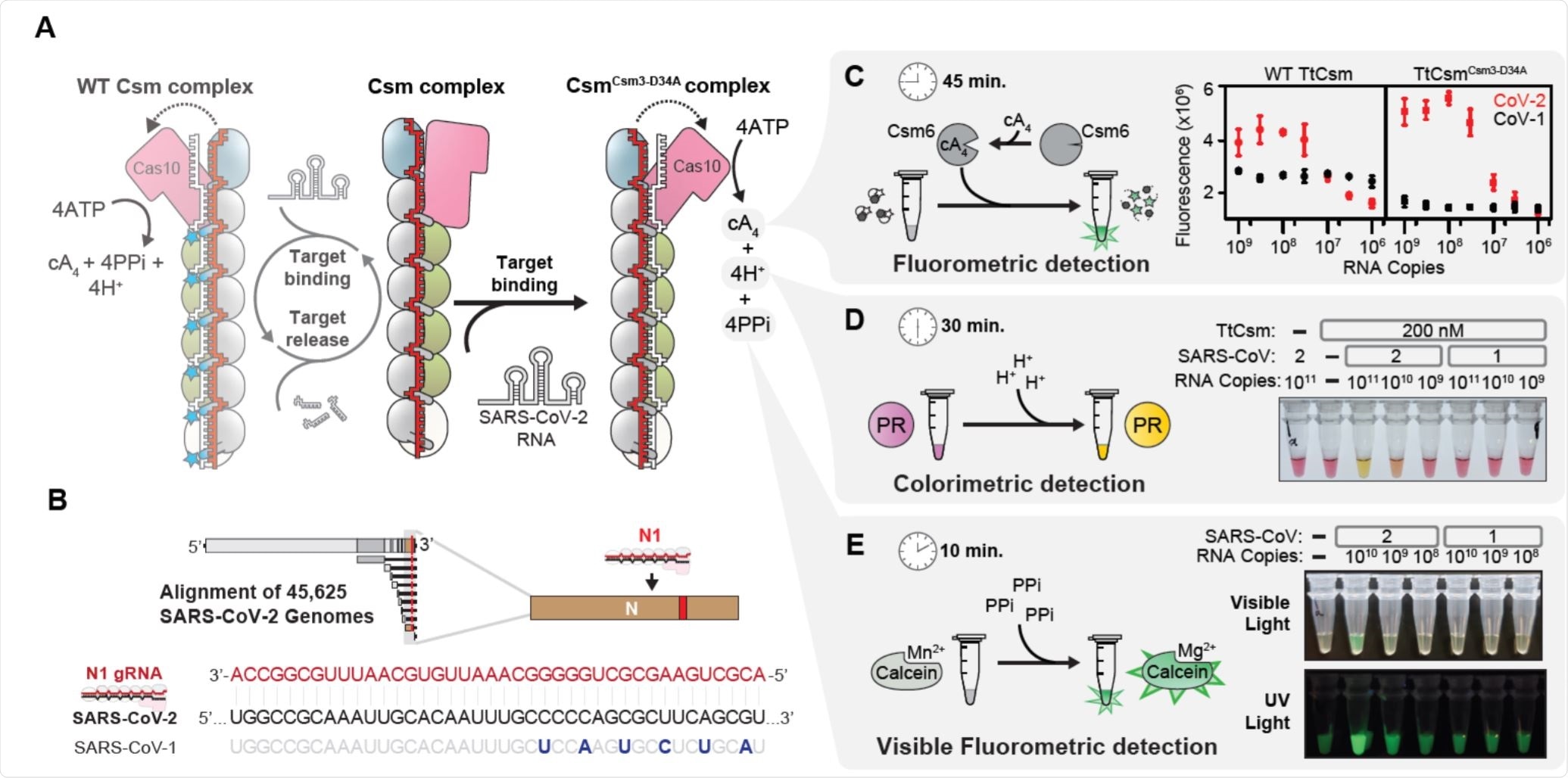
Image Credit:https://www.medrxiv.org/content/10.1101/2020.10.14.20212670v1.full.pdf

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Why use a CRISPR system?
Loop-mediated isothermal amplification (LAMP) is a low-cost DNA amplification technique, producing significantly more DNA than PCR. Type V and IV CRISPR-Cas systems have been utilized in conjunction with LAMP to cleave amplified target RNA or DNA that has been tagged with a fluorophore and quencher, initiating fluorescence and providing a quantitative detection signal.
Combining these systems gives a reliable and sensitive method of identification, though potentially involves a number of amplification steps. CRISPR systems themselves may be used for amplification, and generate a number of detectable side-products in the process.
Type III CRISPR-Cas systems bear the protein Cas10 and can target both DNA and/or RNA, generating a large quantity of crRNA. Pre-crRNA is processed in a sequence-specific manner and trimmed in a ruler-based non-specific way by Cas6. A Csm complex from Thermus thermophilus was programmed to recognize SARS-CoV-2 RNA specifically, activating polymerization by Cas10 and generating a number of by-products including hydrogen protons, cyclic tetra-adenylate, and pyrophosphates, which can be detected by changes in pH or by fluorimetry.
The group selected a target gene seen to be conserved across 99.99% of tested SARS-CoV-2 samples and replaced the CRISPR array of the selected system with a synthetic one that would recognize the target sequence. They also took steps to inactivate Csm3-mediated cleavage in the system, ensuring that the target nucleic acid sequence was only replicated.
Csm3 controls Cas10 polymerase activity to limit nuclease activation, and by inactivating Csm3, Cas10 remains bound to the target RNA for longer. RNA cleavage would lower the concentration of the target RNA over time, lessening the sensitivity of the assay. In a comparative test, the group found that inactivation of Csm3 improved sensitivity by around three times.
LAMP-CRISPR detection of SARS-CoV-2
Sample SARS-CoV-2 RNA was firstly amplified by LAMP, followed by further amplification of target RNA by the CRISPR system, generating sufficient quantities of by-products to be detectable by three unique systems within 10-45 minutes. Compared to reverse transcriptase-quantitative polymerase chain reaction the method is cheap and fast, with less need for sophisticated equipment.
An RNA-tether linking fluorophore and quencher was utilized, which is cleaved by Csm-6 in the presence of cyclic tetra-adenylate, producing a sufficiently fluorescent signal within 45 minutes. The specificity of the system was also demonstrated by comparing with SARS-CoV-1 samples, which generated little fluorescence.
The second system involved the use of pH-sensitive dye Phenol Red, providing a colorimetric indication of the number of protons released during polymerization within 30 minutes.
The third exploited the release of pyrophosphates that disrupt the chelation of a manganese ion to calcein, a fluorophore that is quenched in the presence of manganese ions. This final process was able to give results within just ten minutes and fluorescence is visible to the naked eye or when under a UV-lamp.
Is the system applicable?
SARS-CoV-2 patients capable of spreading the infection generally have a concentration of at least 106 RNA copies per mL of the collected sample. The fluorescent CRISPR system described was able to detect SARS-CoV-2 at concentrations of 107 RNA copies per mL, and so prior amplification by LAMP was still found to be necessary.
The authors hope to improve sensitivity further in the future by improving the homogeneity of the engineered Csm complex they developed, and by screening the produced crRNA for common and unique features that would allow them to be quantified.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
https://www.medrxiv.org/content/10.1101/2020.10.14.20212670v1.full.pdf