Scientists from the University of Groningen, the International Institute of Molecular and Cell Biology in Warsaw, and Leiden University comprehensively studied the RNA genomic structure of the SARS-CoV-2 coronavirus.
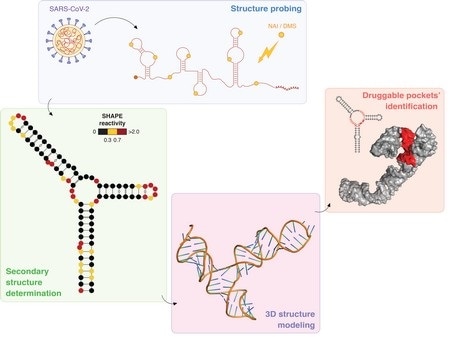
After “probing” the structure of the SARS-CoV-2 RNA genome using specific chemical compounds, the secondary structure of stable RNA elements throughout the viral genome has been determined. Using this information, the 3D model of these RNA structure elements has been computationally determined, allowing the identification of potential pockets that might provide targets for the development of antiviral therapeutic strategies. Image Credit: Danny Incarnato.
These RNA structures are promising targets for developing drugs against the coronavirus. The study results were published as “Breakthrough paper” in the Nucleic Acid Research journal on November 10th, 2020.
Infectious diseases induced by viruses and bacteria have existed all through the history of mankind. But in the last 18 years, a majority of deaths across the world had occurred due to severe acute respiratory syndromes induced by coronaviruses, such as MERS and SARS. This, along with the current COVID-19 pandemic that has already claimed over a million lives, shows that there is an urgent need to develop innovative ways to fight coronavirus infections.
Genome structure
SARS-CoV-2, which causes COVID-19 infection, is a beta coronavirus that has a linear single-stranded, positive-sense RNA genome. Much like those in other RNA viruses, the RNA structures of the SARS-CoV-2 are believed to play a vital role in how this virus replicates in human cells. But in spite of this significance, only a few of functionally pertinent coronavirus structural RNA elements have been examined so far.
Hence, scientists from the IIMCB (Poland), along with researchers from the University of Groningen and Leiden University (both in the Netherlands), conducted a broad characterization of the RNA genome structure of SARS-CoV-2 by employing many sophisticated methods.
Supervised by Dr Danny Incarnato from the University of Groningen, the study involved an analysis of RNA structures to acquire single-base resolution secondary structure maps of the entire genome of SARS-CoV-2 coronavirus in vitro as well as in living infected cells.
The researchers subsequently detected a minimum of 87 regions in the SARS-CoV-2 RNA sequence that seems to form clear compact structures. Among these structures, at least 10% are under robust evolutionary selection pressure between coronaviruses, indicating functional relevance. Most significantly, this is the first study in which the structure of the whole coronavirus RNA (one of the longest viral RNAs around 30,000 nucleotides) was established.
Drug
We first identified the structures in vitro, and subsequently confirmed their presence in the RNA of viruses inside cells. This means that our results are very robust.' Also, pockets were identified in some RNA structures that could be targeted by small molecules to hamper the function of the viral RNA.”
Dr Danny Incarnato, University of Groningen
Dr Incarnato continued, “Furthermore, a number of the structures are conserved between different coronaviruses, meaning that a successful drug targeting SARS-CoV-2 could also be effective against future new virus strains.”
The researchers also detected parts of the SARS-CoV-2 RNA that are innately unstructured. “These could be targeted by antisense oligonucleotide therapeutics,” added Dr Incarnato.
Introducing short nucleic acid strands that can attach to these sections of viral RNA would form double-stranded regions. These regions are naturally targeted by enzymes within the human cells.
Weak spots
To sum up, this collaborative study establishes a solid foundation for upcoming work aimed at designing promising small-molecule drugs for treating SARS-CoV-2 infections and perhaps also infections caused by other coronaviruses.
This work would not have been possible without the collaboration between the Netherlands and Poland. Together, we invented a new way of searching for potential weak spots in large viral RNAs. Collectively, our work lays the foundation for the development of innovative RNA-targeted therapeutic strategies to fight SARS-CoV-2 infections.”
Janusz Bujnicki, Head of the Laboratory of Bioinformatics and Protein Engineering, International Institute of Molecular and Cell Biology
The article explaining the outcomes of these analyses has been published in the Nucleic Acids Research journal, and reviewers who assessed the document nominated it for featured status as a “breakthrough paper”.
Source:
Journal reference:
Manfredonia, I., et al. (2020) Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Research. doi.org/10.1093/nar/gkaa1053.