Researchers from the Tokyo Institute of Technology have developed a mathematical model to quantitatively assess the impacts of particular epigenetic changes on the transcription rates. For this, the team efficiently created reconstituted chromatin-bearing histone modifications in vitro.
Site-specific histone acetylation can differentially affect the sequential stages of transcription. Image Credit: Nucleic Acids Research.
Published in the Nucleic Acids Research journal, the study offers a precise quantitative strategy for interpreting how site-specific modifications to histone proteins influence the accessibility of both chromatin and gene expression levels.
The development of a single protein is a long and complex procedure. Even the creation of its own blueprint, called a coding transcript, from a DNA template involves multiple players and steps.
To begin with, the DNA is generally found neatly enclosed around proteins, known as histones, to create a nucleosome, the basic subunit of a closely condensed structure, known as chromatin. The degree of its condensation decides the amount of DNA that is “available” for the transcription process.
Histone modifications, like acetylation, also affect the accessibility of chromatin for gene expression. Such “epigenetic” changes play a vital function in controlling the expression of genes. So far, not much has been studied regarding these epigenetic effects, how they can be precisely quantified, and how site-specific histone modifications can have an impact on gene expression.
To address these questions, a research team headed by Professor Masahiro Takinoue from the Tokyo Institute of Technology, Professor Kohki Okabe from The University of Tokyo, and Professor Takashi Umehara from the RIKEN Center for Biosystems Dynamics Research in Japan has established a kinetic model to measure the contribution of epigenetic modifications on the rate of transcription based on highly quantitative experimental outcomes.
Referring to their latest study published in the Nucleic Acids Research journal, Takinoue explained, “The contribution of each modification state of each histone to the sequential steps of chromatin transcription was yet to be quantified because of the difficulty in precise reconstitution of a chromatin template with the epigenetic modification(s) of interest and quantification of RNA transcription from it.”
Using genetic code expansion and cell-free protein synthesis, we synthesized histone H4 containing designed site-specific acetylation(s) and reconstituted a tetra-acetylated nucleosome.”
Masahiro Takinoue, Professor, Tokyo Institute of Technology
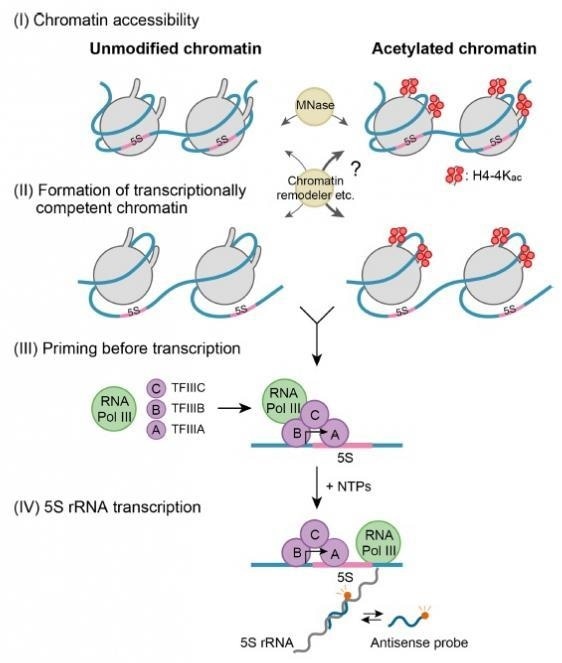
Histone modifications focus on different stages in the transcription process at the chromatin level, which could consequently affect the transcription rate. The initial stage, called “chromatin accessibility,” involves the opening up of the closely condensed structure and rendering it accessible to the transcription machinery.
The second phase, called the “formation of transcriptionally competent chromatin,” prepares the chromatin to enable the attachment of transcript synthesizing complexes. The third stage, called “priming before transcription,” supports the assembly of accessory proteins needed to initiate transcription. In the end stage of transcription, nucleotides are sequentially added to form the transcript.
To analyze how site-specific histone modifications, such as acetylation, influence the transcription stages, the team produced a reconstituted nucleosome that carries two copies of RNA coding sequence from a frog species, called Xenopus, with acetylation on certain sites of the histone proteins.
Such a system replicates the dynamic chromatin changes inside the living cell, which influence transcription. The team also created a precise and highly responsive fluorescence-based detection device that can quantify the trace concentrations of transcripts in real time.
When the researchers applied their kinetic model, they observed that acetylation at four particular histone sites enhances the accessibility of chromatin by three times when compared to chromatin that lacks acetylation.
Moreover, the versatility of the model enables it to be utilized for measuring other epigenetic changes. Emphasizing the promising applications of their research work, Takinoue concluded, “Our mathematically described kinetic model allowed us. Our methodology will be applicable to a wide variety of chromatin-mediated reactions for quantitative understanding of the importance of epigenetic modifications.”
Source:
Journal reference:
Wakamori, M., et al. (2020) Quantification of the effect of site-specific histone acetylation on chromatin transcription rate. Nucleic Acids Research. doi.org/10.1093/nar/gkaa1050.