A collaborative team of researchers from The Korea Advanced Institute of Science and Technology (KAIST) and the University of California San Diego (UCSD) has developed a deep neural network called DeepTFactor that predicts transcription factors from protein sequences.
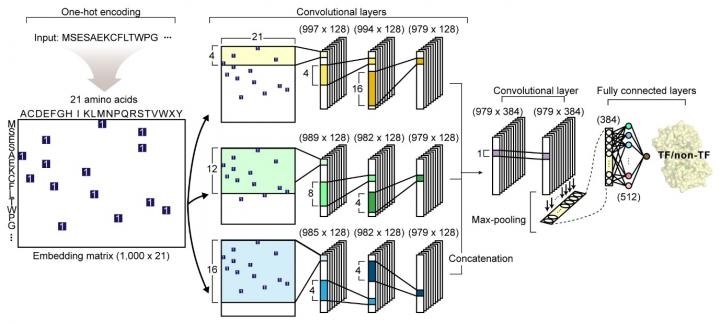
The network architecture of DeepTFactor. An input protein sequence is processed using three parallel subnetworks. Image Credit: KAIST.
The DeepTFactor acts as a useful tool for knowing the regulatory systems of organisms and expediting the use of deep learning to find solutions to biological problems.
A transcription factor is a protein that binds particularly to DNA sequences and controls the initiation of transcription. The analysis of transcriptional regulation helps to understand how organisms regulate gene expression in response to environmental or genetic changes. To this end, the first step in analyzing the transcriptional regulatory system of an organism is to find the transcription factor of an organism.
In previous studies, transcription factors have been predicted through the analysis of sequence homology with the already characterized transcription factors or by data-driven methods like machine learning.
Traditional machine learning models necessitate a rigorous feature selection method that depends on domain expertise like estimating the physicochemical properties of molecules or investigating the homology of biological sequences. At the same time, deep learning can intrinsically study the latent features for a particular task.
The collaborative research team includes PhD candidate Gi Bae Kim and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering at KAIST, and Ye Gao and Professor Bernhard O. Palsson from the Department of Biochemical Engineering at UCSD.
The study titled “DeepTFactor: A deep learning-based tool for the prediction of transcription factors” was published online in PNAS. The study describes the development of DeepTFactor—a deep learning-based tool that uses three parallel convolutional neural networks to predict whether a given protein sequence is a transcription factor.
The researchers predicted 332 transcription factors of Escherichia coli K-12 MG1655 using DeepTFactor and the operation of DeepTFactor by experimentally validating the genome-wide binding sites of three predicted transcription factors (YqhC, YiaU, and YahB).
Furthermore, the team employed a saliency approach to gain insights into the reasoning process behind DeepTFactor. They confirmed that though the information on the DNA binding domains of the transcription factor was not explicit from the training process, DeepTFactor implicitly learned and used it for the prediction.
In contrast to earlier tools for predicting transcription factor—that were developed only for protein sequences of specific organisms—it is expected that DeepTFactor could be used to analyze the transcription systems of all organisms at a high level of performance.
DeepTFactor can be used to discover unknown transcription factors from numerous protein sequences that have not yet been characterized. It is expected that DeepTFactor will serve as an important tool for analyzing the regulatory systems of organisms of interest.”
Sang Yup Lee, Distinguished Professor, Department of Chemical and Biomolecular Engineering, KAIST
Source:
Journal reference:
Kim, G. B., et al. (2021) DeepTFactor: A deep learning-based tool for the prediction of transcription factors. Proceedings of the National Academy of Sciences. doi.org/10.1073/pnas.2021171118.