If finding a needle in a haystack is hard enough, then one should try finding a particular molecule on the needle.
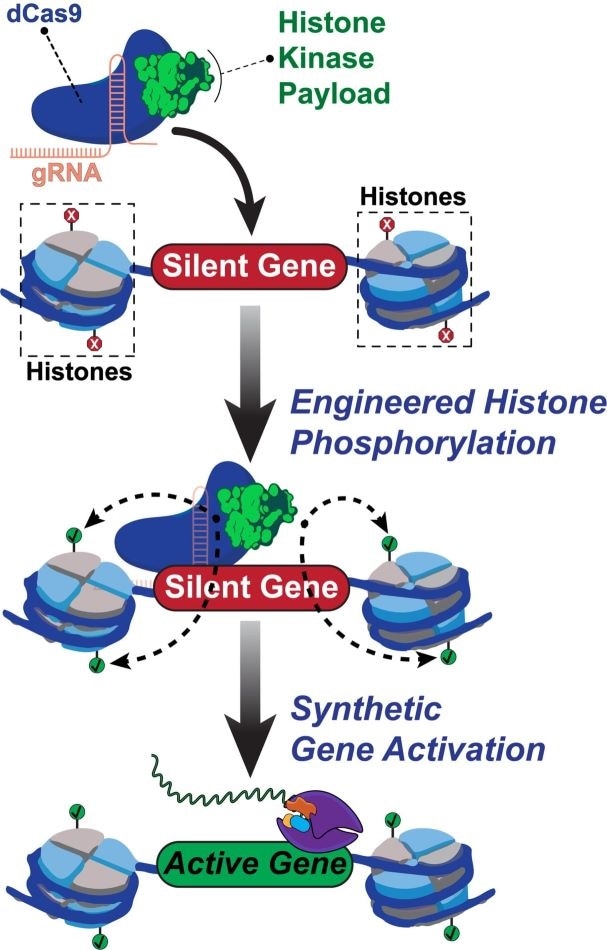
Rice University scientists built a new tool to engineer and understand how human genes are turned on. The team created a synthetic two-part protein based on dCas9 and a modified enzyme called dMSK1 to deliver chemical payloads at precise spots near human genes. The tool causes pinpoint changes to histone marks and with the help of other proteins, the activation of silent human genes. Image Credit: Hilton Laboratory.
Scientists from Rice University have accomplished something along these lines with a novel genome-editing tool that focuses on the supporting players involved in a cell’s nucleus that not only package DNA but also help in the expression of genes. The new study paves the way to new treatments for various diseases, including cancer.
Isaac Hilton, a bioengineer from Rice University, and Jing Li, a postdoctoral researcher and lead author of the study, and their collaborators programmed an altered CRISPR/Cas9 complex to target particular histones, which are universal epigenetic proteins that maintain the DNA order, with pinpoint precision.
The open-access article was published in the Nature Communications journal.
Histones help control several cellular processes. Each nucleosome (the basic “beads on a string” in DNA) contains four histones that help regulate the function and structure of human genomes by revealing genes for activation.
According to Hilton, “Nucleosomes serve as architectural substrates to fit our DNA inside of our cells, and can also control access to key parts of our genomes.”
Histones, which are similar to other proteins, can be activated by phosphorylation—a biochemical process in which a phosphoryl group is added to regulate protein-DNA or protein-protein interactions.
Histones can display an exquisitely diverse spectrum of chemical modifications that serve as beacons or regulatory markers and tell which genes to turn on, and when, and how much to do so. One of these mysterious modifications is phosphorylation, and we aimed to better illuminate the mechanism by which it can rapidly turn human genes on and off.”
Isaac Hilton, Bioengineer, Rice University
According to him, no other epigenome editing method has allowed site-specific control across the phosphorylation of histones. Developed by Rice University, the programmable tool known as dCas9-dMSK1 combines a “hyperactive” human histone kinase—an enzyme that catalyzes phosphorylation—and a deactivated “dCas9” protein.
The CRISPR/Cas9 complex generally uses Cas9 “scissors” and guide RNAs to target and cut DNA sequences. The novel tool programs the deactivated dCas9 protein to target without cutting sequences and instead use the recruited dMSK1 enzyme to phosphorylate the histone of interest and switch on proximal genes.
The team used the dCas9-dMSK1 tool to reveal new genes and pathways that are essential for drug resistance. Li used the tool to detect three genes that were formerly associated with melanoma drug resistance earlier.
And then she identified seven new genes linked to melanoma resistance. It's an exciting finding that we are following up on. Histone proteins that wrap up DNA can have all sorts of chemical marks and combinations on them. This results in what has been dubbed a histone code, and one of our goals is to work to decipher it.”
Isaac Hilton, Bioengineer, Rice University
Li’s tool also demonstrated how particular histone marks interact with one another.
It tells us that chemical modifications on histones talk to each other, and we can show it happening at specific spots in the human genome. And that's linked to a gene turning on, so this allows us to synthetically control them.”
Jing Li, Study Lead Author, Rice University
Li added that a long-term objective is to target a wide range of other histone marks. It's a complicated story. There are a lot of different positions and features of histones that we want to study,” she said.
“Getting these technologies into patients is a long process. But tools like this are the first step and can pave the way towards understanding how normal cellular processes, unfortunately, go awry in human diseases,” Hilton concluded.
Source:
Journal reference:
Li, J., et al. (2021) Programmable human histone phosphorylation and gene activation using a CRISPR/Cas9-based chromatin kinase. Nature Communications. doi.org/10.1038/s41467-021-21188-2.