A new tool developed by a multidisciplinary team of mathematicians and biologists from the University of California, Irvine could help decode the language used by cells to interact with each other.
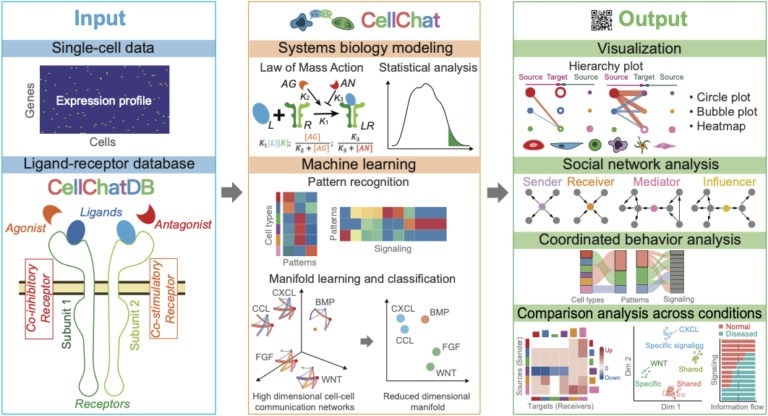
Overview of how CellChat can convert "molecular language" of cells into the translation that is interpretable by researchers. Image Credit: Suoqin Jin, Qing Nie & Maksim Plikus / UCI.
In a new study recently published in the Nature Communications journal, the research team introduced a new computational platform—called CellChat—that helps decode the signaling molecules that convey commands and information between the cells that come collectively to form biological tissues and even whole organs.
To properly understand why cells do certain things, and to predict their future actions, we need to be able to listen to what they are saying to one another; mathematical and machine learning tools enable the translation of such messages.”
Qing Nie, Study Co-Senior Author and Professor, Mathematics and Developmental & Cell Biology, University of California, Irvine
Qing Nie is also the Chancellor at the University of California, Irvine.
“Just like in our world, where we are constantly bombarded with information, all cells experience a lot of molecular words coming at them simultaneously. What they choose to do is dependent on this steady flow of molecular information and on what words and sentences are being heard the loudest,” stated Maksim Plikus, the co-senior author of the study and professor of Developmental & Cell Biology at the University of California, Irvine.
To use the CellChat platform to interpret the molecular messages between cells, the team fed in a single-cell gene expression and this yielded a comprehensive report on the signaling communication traits of a specified organ or tissue.
For each distinct group of cells, CellChat shows what significant signals are being sent to their neighbors and what signals they have the ability to receive. As an interpreter of cellular language, CellChat provides scientists with a valuable insight into signaling patterns that guide function of the entire organ.”
Maksim Plikus, Study Co-Senior Author and Professor, Developmental & Cell Biology, University of California, Irvine
For designing the CellChat platform, the scientists from the NSF-Simons Center for Multiscale Cell Fate Research at the University of California, Irvine—including postdoctoral fellows Suoqin Jin, Christian F. Guerrero-Juarez, Raul Ramos, and Lihua Zhang, borrowed heavily from social network theory and machine learning tools, which enables the CellChat platform to forecast higher-level meaning of cellular language and recognize contextual similarities that are otherwise not evident. The platform breaks down the enormous complexity of the patterns of cellular communication.
Cells generate modifier molecules to highlight a specific command, translating “do this” to “do this now.” With the CellChat platform, the strength of the signaling capacity of cell communication is automatically calibrated by taking into account all the substantially existing modifier molecules.
Therefore, its translation turns out to be more nuanced and helps reduce errors that are common in other analogous yet less advanced computational tools.
Beyond the sole underlying research enterprise of understanding these biological messages, the CellChat platform can also be utilized to compare communication networks in various states of an organ, like health and sickness, Nie added.
Dubbing it a “Google Translator for the lexicon of cells,” Nie added that one of the most important capabilities of the tool is that it can be used to reveal molecular drivers in a broad spectrum of diseases, such as autoimmune disorders and cancer.
In our paper, we showcase the power of CellChat using atopic dermatitis, a human skin condition, but the tool can be used on any tissue with the same success.”
Maksim Plikus, Study Co-Senior Author and Professor, Developmental & Cell Biology, University of California, Irvine
Source:
Journal reference:
Jin, S., et al. (2021) Inference and analysis of cell-cell communication using CellChat. Nature Communications. doi.org/10.1038/s41467-021-21246-9.