In an effort to contribute to the fight against cancer, Swedish researchers have published novel molecular mapping of proteins that controls the process of cell division—detecting 300 of these proteins.
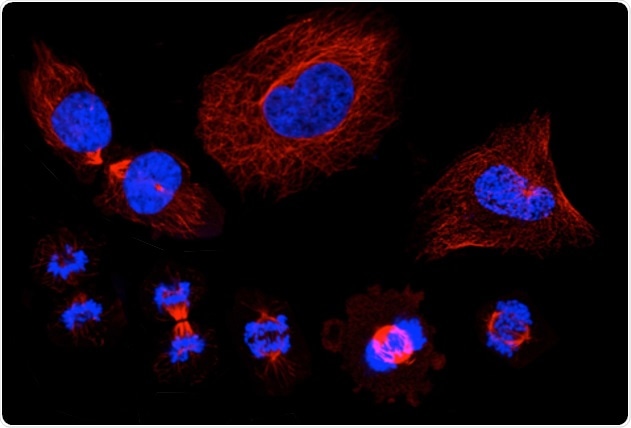
A montage of microscopic images that show the stages of the cell cycle, in which a single cell duplicates its DNA and eventually divides into two "daughter" cells. Staining shows the cell nucleus in blue, and the microtubules in red. Image Credit: KTH Royal Institute of Technology/SciLifeLab.
The study results, which were recently published in Nature—a scientific journal—are important because they help bring medical research closer to the point of being able to focus on certain proteins to treat cancer.
According to Emma Lundberg, the co-author of the study and a professor from KTH Royal Institute of Technology, it is important to identify and interpret the factors that define these proteins, Lundberg’s research team at the Science for Life Laboratory (SciLifeLab) in Stockholm contributed to the plotting of these proteins.
The long-term goal is such protein mapping will lead to the development of personalized cancer medications and therapies, which are adapted to the particular anatomical state of the individual patient relating to the underlying disease, added Lundberg.
Apart from the 300 newly-discovered proteins, the team reported that 20% of the human proteome (all protein molecules encoded by the genome) indicates intra-cellular variations, that is, variations in gene expression within the otherwise matching cells.
This data provides medical research with novel insights into the cell cycle, in which an equilibrium is moderated between certain proteins that support the proliferation of cells and those that suppress it. According to Lundberg, the study is now integrated into the Human Protein Atlas—an open-access research database.
Our hope is that this provides a valuable resource for a better understanding of, among other things: cell-to-cell variation, the human cell cycle, and the newly-identified proteins in the cell cycle and their role in the formation of tumors.”
Emma Lundberg, Professor, KTH Royal Institute of Technology
The researchers applied the so-called immunofluorescence microscopy to detect proteins that are specific to the cell cycle and then merged the collected data with the RNA sequencing of separate cells to explain the temporal existence of proteins and RNA throughout the cell cycle.
Source:
Journal reference:
Mahdessian, D., et al. (2021) Spatiotemporal dissection of the cell cycle with single-cell proteogenomics. Nature. doi.org/10.1038/s41586-021-03232-9.