Precisely two decades ago after the successful conclusion of the “Human Genome Project”, an international research team, the Human Genome Structural Variation Consortium (HGSVC), with the contribution of Heinrich Heine University Düsseldorf (HHU), has currently sequenced a total of 64 human genomes at high resolution.
Comprehensive discovery of genetic variation based on analysis of human genomes of diverse ancestry. Image Credit: David Porubsky, University of Washington.
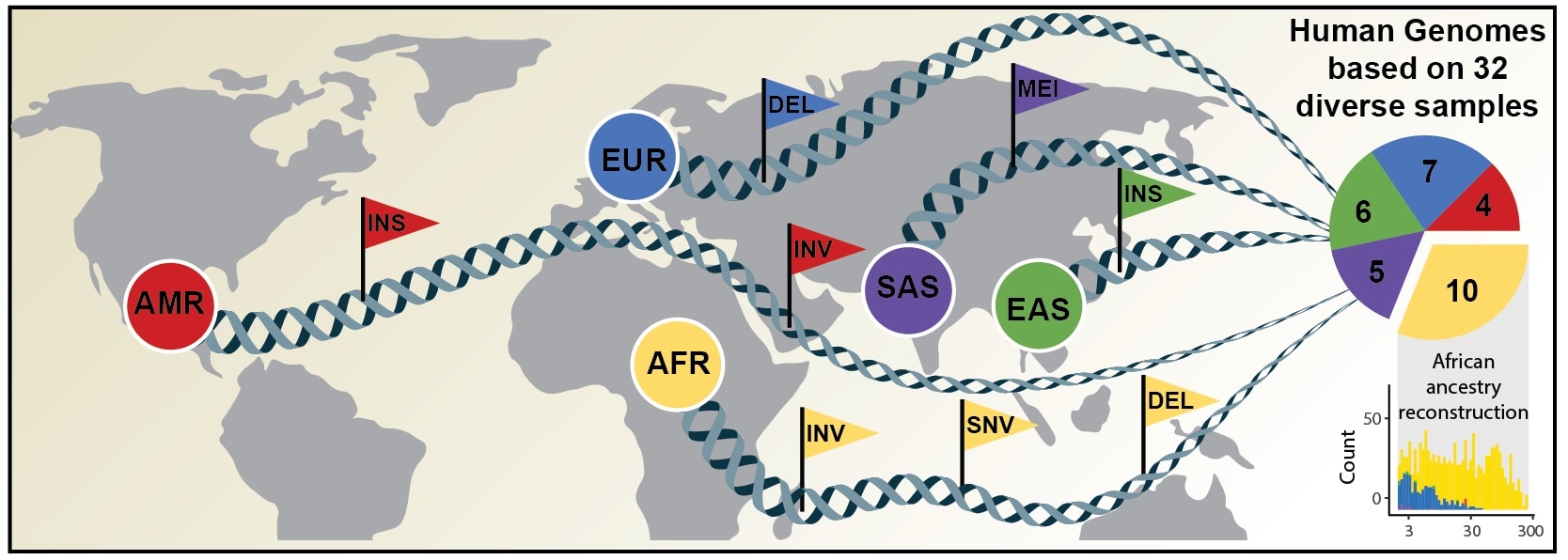
This reference data comprises individuals from across the world, better recording the different types of genes in the human species. Among other applications, the study allows population-specific analyses on genetic predispositions to various human diseases and also the finding of more complex forms of genetic variation, as reported by 65 authors in the latest issue of the scientific journal, Science.
Back in 2001, the “International Human Genome Sequencing Consortium” declared the first draft of the human genome reference sequence to the world. The so-called Human Genome Project had taken over 11 years of work and included over 1000 researchers from 40 nations. But this reference data did not represent even a single individual but was rather a composite of humans that could not precisely capture the difficulty of the variations of human genes.
Based on this, researchers have performed several sequencing studies over the last two decades to detect and catalog genetic variations between the reference genome and an individual. Such variations generally targeted tiny single base changes and overlooked greater genetic changes.
Now, existing technologies have started to identify and define bigger variations—known as structural variants—like insertions of many hundred letters. Compared to smaller genetic variations, structural variants are more likely to disrupt the function of genes.
Now, an international group of scientists has published an article in the Science journal, declaring a new and significantly more extensive reference dataset achieved using a mix of sophisticated sequencing and mapping technologies.
The latest reference dataset reflects a total of 64 assembled human genomes, denoting 25 various human populations worldwide. Most significantly, all genomes were organized without direction from the initial human genome and thus better captures genetic variations from different populations of human beings.
The new analysis was headed by researchers from the European Molecular Biology Laboratory Heidelberg (EMBL), the Heinrich Heine University Düsseldorf (HHU), The Jackson Laboratory for Genomic Medicine in Farmington, Connecticut. (JAX), and the University of Washington in Seattle (UW).
With these new reference data, genetic differences can be studied with unprecedented accuracy against the background of global genetic variation, which facilitates the biomedical evaluation of genetic variants carried by an individual.”
Dr Peter Ebert, Study Co-First Author, Institute of Medical Biometry and Bioinformatics, Heinrich Heine University
Due to spontaneous and constantly occurring changes in the genetic material, the distribution of genetic variants can considerably vary between the population groups. If such a mutation is transferred across several generations, it can turn out to be a genetic variant that is specific to that population.
The latest reference data offers a crucial basis for including the entire spectrum of genetic variants in the supposed genome-wide association analyses. The objective is to predict the individual risk of developing specific diseases, like cancer, and to interpret the fundamental molecular mechanisms. This, consequently, can be utilized as a basis for additional targeted therapies and preventative medicine.
The study may allow more applications in precision medicine. For instance, drug efficacy can differ between individuals on the basis of their genomes. Now, the latest reference data represents the complete range of diverse types of genetic variants and combines human genomes of great diversity.
Hence, this new resource might help develop innovative methods in personalized medicine, in which the choice of treatments is adapted to the individual genetic background of the patient.
Just a few years ago, I would not have imagined that resolving genomes to this completeness would become possible so fast. This was enabled by exciting advances both of biotechnological and computational methods. Great to see this technology applied to a diversity panel of human genomes. These genome sequences will be an important resource for fundamental research and clinical genomics going forward.”
Dr Peter Ebert, Study Co-First Author and Computational Biologist, Heinrich Heine University Düsseldorf
Dr. Tobias Marschall, the senior author of the study and who headed the study at HHU, added, “It was especially exciting to see that these new genome sequences enable a much more detailed analysis of data from standard sequencing technologies, which are routinely applied to millions of genomes by researchers and clinicians across the globe."
Dr. Marschall believes that “future studies to find associations between genetic variants and disease susceptibility will clearly benefit from this new approach.”
The research builds on a new approach published by the team in 2020 in the Nature Biotechnology journal to precisely rebuild the two parts of an individual’s genome—one inherited from a person’s mother and the other inherited from the father.
When arranging the genome of a specific person, this technique removes the potential biases that may result from comparisons with a defective reference genome.
Dr. Ebert emphasized the interdisciplinary cooperation at HHU, “We performed our extensive computations on the High-Performance Computing Cluster Hilbert. The HPC team of the Düsseldorf ZIM thus had an important role for the success of our research project.”
Source:
Journal reference:
Ebert, P., et al. (2021) Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. doi.org/10.1126/science.abf7117.