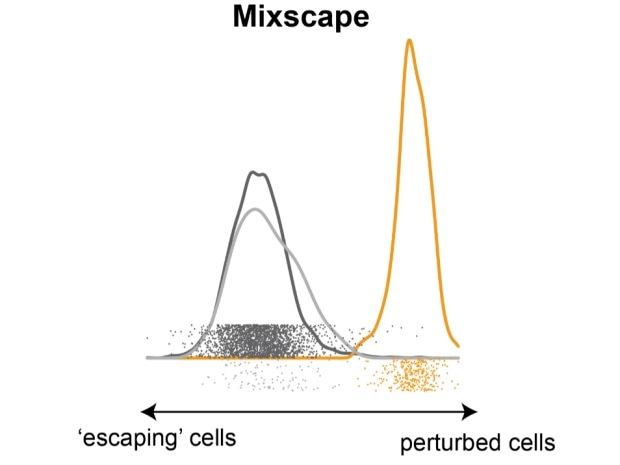
Mixscape separates a mixture of cells into perturbed (yellow) and non-perturbed (‘escaping,’ grey) cells. Image Credit: New York University. New York Genome Center
The new article explains how the innovative technique, known as mixscape, helped to detect a novel molecular mechanism for regulating immune checkpoint proteins that control the ability of the immune system to detect and kill tumor cells.
Our approach will help scientists to connect genes to the specific cellular behaviors and molecular pathways that they regulate.”
Rahul Satija, Study Senior Author and Associate Professor of Biology, Center for Genomics and Systems Biology, New York University
Satija is also a core faculty member at the New York Genome Center.
The team set out to gain a better understanding of how tumor cells modify the regulation of the crucial gene, for example, the immune checkpoint molecule, called PD-L1 to avoid detection and elude the body’s immune system.
To achieve this, the researchers conducted a pooled genetic screen, in which they “knocked out” a group of genes in a cancer cell line model to note the effect of each change perturbation or change on PD-L1 levels.
The team used a technology, called ECCITE-seq, that enables scientists to capture single-cell profiles of varied types of biomolecules—like protein and RNA—after perturbing every gene with a CRISPR “guide RNA.”
The potential to quantify numerous types of molecular data, called multimodal analysis, enabled the researchers to differentiate between post-transcriptional and transcriptional modes of regulation.
But after finishing the experiments, the researchers realized that important computational challenges restricted its potential to examine and understand the data. For instance, the team discovered that when the same gene is knocked out in various different cells, the results had a striking variability.
Specifically, a considerable fraction of cells—up to 75% in certain cases—seemed up to escape any discernible effects after the attempted perturbation and denoted a perplexing source of noise in downstream analysis.
Facing these challenges made us realize that we needed new computational methods to identify and remove confounding sources of variation in our dataset.”
Efthymia Papalexi, Study Lead Author and Biology Graduate Student, New York University
To accomplish this, the researchers created a statistical method—mixscape—to model every perturbation so as to give rise to a combination of cells with various reactions. While doing so, the mixscape technique can detect and eliminate sources of noise from the data, thus enabling the user to target the most significant biological signals that persist.
When we applied mixscape in our screen, we boosted our power to connect gene perturbations with changes in the transcriptome and protein expression. This allowed us to discover that the kelch-like protein KEAP1 and the transcriptional activator NRF2 mediate a cell’s expression level of PD-L1.”
Rahul Satija, Study Senior Author and Associate Professor of Biology, Center for Genomics and Systems Biology, New York University
Although these analyses were performed in cancer cell lines, NRF2 and KEAP1 are often mutated in samples of human lung cancer, indicating that these genes may significantly contribute to the development and progression of tumors in humans.
Looking ahead, the team is exploiting mixscape and multimodal single-cell pooled CRISPR screens to interpret the molecular regulation of scores of more pathways and cellular behaviors.
Mixscape is now freely available online via the Satija laboratory’s Seurat package—a software toolkit meant for biomedical scientists.
“We hope our method will be useful for the community and assist in the study of how genes and molecular pathways interact with each other,” concluded Papalexi.
Source:
Journal reference:
Papalexi, E., et al. (2021) Characterizing the molecular regulation of inhibitory immune checkpoints with multimodal single-cell screens. Nature Genetics. doi.org/10.1038/s41588-021-00778-2.