Back in 1993, researchers had discovered that Huntington’s disease is typically caused by a single mutated gene, known as HTT, creating high expectations for a rapid cure. But even today, no approved treatment is available for this disease.
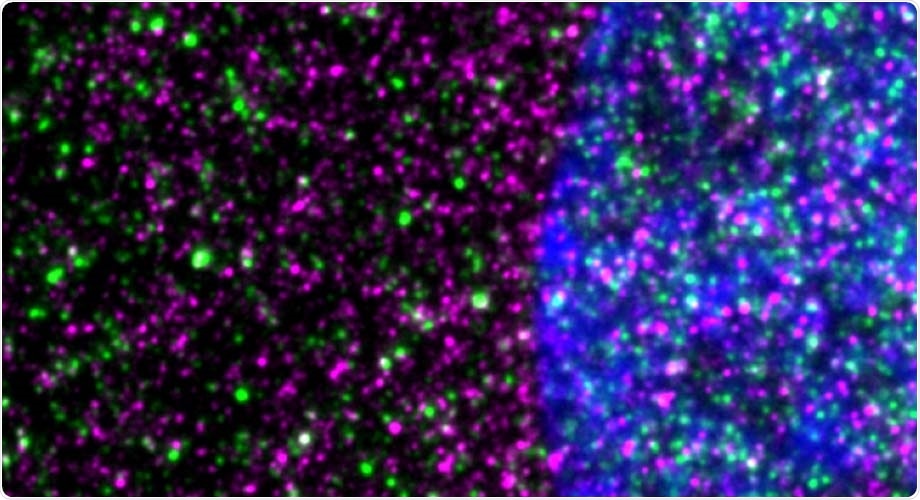
Disease-causing huntingtin, shown in red, interacts with ribosomes, shown in green, in a striatal neuron. The nucleus is blue. Image Credit: Nicolai Urban of Max Planck Institute for Neuroscience in Jupiter, Florida.
According to neuroscientist Srinivasa Subramaniam, Ph.D., from the Scripps Research Institute, one challenge has been limited knowledge of how the mutant huntingtin protein triggers the death of brain cells. Now, Subramaniam’s team has demonstrated that the mutated huntingtin protein delays the protein-building machines, known as ribosomes, in the brain cells. This latest study was published in the Nature Communications journal.
The ribosome has to keep moving along to build the proteins, but in Huntington’s disease, the ribosome is slowed. The difference may be two, three, four-fold slower. That makes all the difference.”
Srinivasa Subramaniam, PhD, Neuroscientist, Scripps Research Institute
There are millions of ribosomes in cells, all circulating along and assembling proteins and amino acids by using genetic information. According to Subramaniam, impairment of the ribosome function eventually has a major impact on the cell.
It’s not possible for the cell to stay alive without protein production.”
Srinivasa Subramaniam, PhD, Neuroscientist, Scripps Research Institute
Subramaniam added that recent advances in gene-translation tracking technologies had enabled the researchers to make these discoveries. These results point to a new way for therapeutic advancements and hold implications for a variety of neurodegenerative diseases, where ribosome stalling seems to play a key role.
In the United States, approximately 10 individuals per 100,000 are affected by Huntington’s disease. This disorder is induced by a surplus number of genetic copies of three DNA-building blocks. Popularly known by the letters CAG, which stands for cytosine, adenine, and guanine, 40 or more of these genetic repeats in the HTT gene lead to brain degenerative disease, which is eventually fatal.
The presence of more repeats leads to the earlier onset of symptoms, which is characterized by behavioral disturbances, balance and movement difficulties, difficulty in eating and speaking, and weakness. These symptoms are caused by brain tissue degeneration that starts in the region known as the striatum and then proliferates. This region is found deep in the brain’s center that regulates voluntary activity and reacts to social reward.
For their studies, the researchers used striatal cells engineered with three varying degrees of CAG repeats in the HTT gene. They studied the effect of the CAG repeats through an advanced technology, known as high-resolution global ribosome footprint profiling (Ribo-Seq), and also the mRNA-seq technique, which provides a snapshot of which genes are active and which are inactive in a given cell at a given time).
The team observed that the translation of many, but not all, proteins were delayed in Huntington’s cells. To validate this discovery, they inhibited the ability of the cells to produce mutant huntingtin protein and observed an increase in the speed of protein synthesis and ribosome movement.
The researchers also assessed how the mutant huntingtin protein affected the translation of other kinds of genes and ruled out the possibility that the slowing effect is probably caused by another ribosome-binding protein, called Fmrp.
Additional experiments provided some signs as to how the mutant huntingtin protein disrupted the function of the ribosomes. They discovered that this protein is directly attached to ribosomal assembly and ribosomal proteins, affecting both the speed of protein synthesis speed and the ribosomal density inside the cell.
According to Subramaniam, although many questions continue to remain, this breakthrough provides a new direction for supporting people with Huntington’s disease.
The idea that the ribosome can stall before a CAG repeat is something people have predicted. We can show that it’s there. There’s a lot of additional work that needs to be done to figure out how the CAG repeat stalls the ribosome, and then perhaps we can make medications to counteract it.”
Srinivasa Subramaniam, PhD, Neuroscientist, Scripps Research Institute
Source:
Journal reference:
Eshraghi, M., et al. (2021) Mutant Huntingtin stalls ribosomes and represses protein synthesis in a cellular model of Huntington disease. Nature Communications. doi.org/10.1038/s41467-021-21637-y.