During biomarker testing, certain disease-associated molecules are examined to estimate the progression of a disease and its response to treatments; however, its application has made it difficult to diagnose non-small-cell lung cancer (NSCLC).
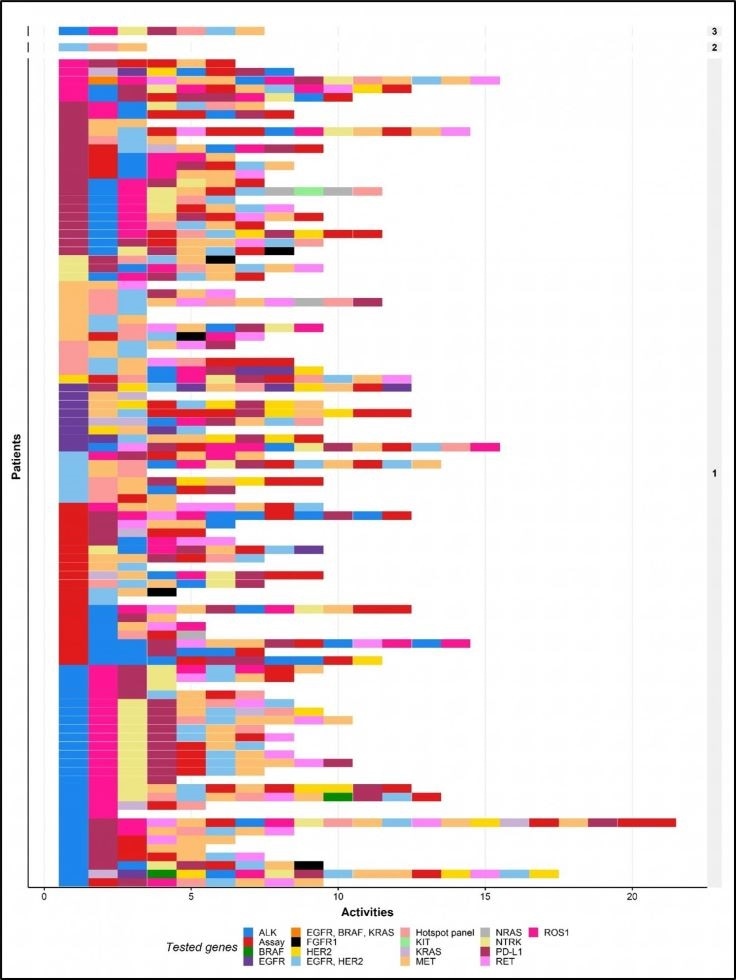
Unique biomarker-test combinations for all individual patients included in the patient cohort. The tests are ordered chronologically. Each row represents the biomarker test combination for one patient. The numbers shown on the right indicate the number of patients who received the same biomarker test combination. Image Credit: Michiel van de Ven., et al.
Now, for the first time, researchers have provided a comprehensive overview of biomarker testing, covering multiple lines of treatments, in a single cohort of patients. The study was published in Elsevier’s The Journal of Molecular Diagnosis.
With the help of exploratory data analysis and process-mining methods in a real-world environment, the team discovered a substantial difference in the use and treatment of a test.
The researchers also discovered that while the whole-genome sequencing technique, in which the unique DNA of a patient is mapped immediately, may not offer a cost-effective option to biomarker testing, as suggested by certain researchers, it may have other advantages for patients, like reducing the time involved between testing and treatment.
While there is a lot of interest in the use of real-world data to analyze treatment variation and outcomes, this study demonstrates the importance of identifying variation in the use of molecular tests as the gateway to most cancer treatments.”
Maarten IJzerman, Professor, Health Technology and Services Research, TechMed Center, University of Twente
IJzerman is also the principal investigator of the study and works at the University of Melbourne Centre for Cancer Research in Melbourne, Australia.
The researchers had access to the entire biomarker testing history of 102 patients suffering from stage IV NSCLC and who had been referred to an all-inclusive cancer center in the Netherlands. When patients have a complicated case, or enrolled in a clinical trial, or have exhausted all treatment options at the referring hospital, they are generally referred to such a facility.
Linked pathology data from the referring hospitals is used to identify eligible patients to ensure the analysis of their comprehensive diagnostic pathways. Such process-mining techniques help identify the actual ordering of treatment procedures and assess the characteristics of testing, like costs and turnaround times.
This is the first-ever study to offer a detailed report containing tested genes, costs, utilized techniques, and turnaround times on the whole sequence of tests administered to patients with stage IV NSCLC.
The team identified 99 unique biomarker-test combinations in 102 patients; practically none of the patients underwent the same kinds of tests in the same order. The most standard biomarkers were generally tested in the first several tests, while emerging biomarkers were usually tested afterward.
The number of tests performed on each patient demonstrated a significant variation. For each patient, tests were performed at different times, and in the majority of the patients, more than one test was performed.
The mean cost of biomarker testing for each patient was US$2258.52/€1881.23. Based on the assumed expense, the number of patients for whom biomarker testing would be equally or less costly if their whole test sequence substituted by whole-genome sequencing ranged between 2 and 29.
We have shown that it is currently unlikely that replacing the current practice of molecular testing in lung cancer with whole-genome sequencing will be cost-saving. Yet, for whole-genome sequencing to be routinely used there are other aspects where it adds value, such as the discovery of other driver mutations not routinely tested for and the potential to streamline diagnostic workflows.”
Michiel van de Ven, Study First Author and PhD Candidate, TechMed Center, University of Twente, Enschede
Numerous biomarker tests for NSCLC may lead to unwanted treatment delays. The large difference in biomarker tests among patients indicates the possibility of inequalities in access to the tests and, thus, access to immunotherapy or targeted treatment.
Whole-genome sequencing may reduce the complexity of the diagnostic pathway, but it has not yet been explored in detail. It could be an exciting avenue for future research.”
Maarten IJzerman, Professor, Health Technology and Services Research, TechMed Center, University of Twente
Source:
Journal reference:
van de Ven, M., et al. (2021) Real-World Utilization of Biomarker Testing for Patients with Advanced Non–Small Cell Lung Cancer in a Tertiary Referral Center and Referring Hospitals. The Journal of Molecular Diagnosis. doi.org/10.1016/j.jmoldx.2021.01.004.