Scientists from the Oregon State University College of Science have achieved a huge step toward new vaccines and drugs for fighting against COVID-19 with an in-depth investigation of the interactions of one protein with SARS-CoV-2 genetic material.
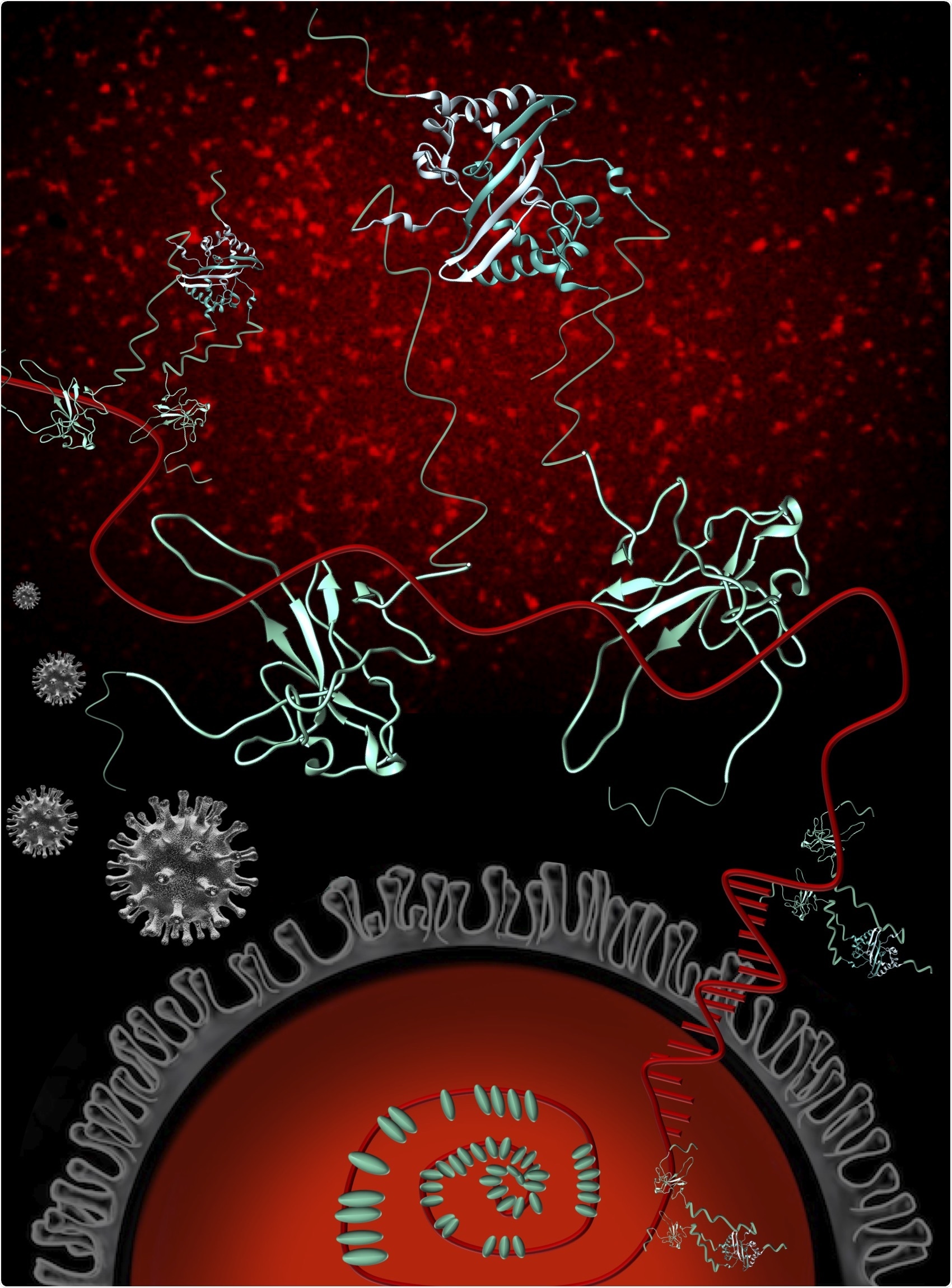
The nucleocapsid phosphoprotein (blue) of SARS-CoV-2 (N) (grey) plays critical roles in multiple processes of the SARS-CoV-2 infection cycle, including replication and transcription, and packaging and protecting the genomic RNA (gRNA) (red). The N protein exists as a dimer in solution and interacts with gRNA predominantly through its structured N-terminal domain. N binds RNA multivalently and as more N proteins become available, stabilizing interactions between RNA and proteins occur, resulting in an organized nucleocapsid. Fluorescence imaging of 1-1000 RNA with a Cy3 fluorescent tag demonstrates that RNA-Cy3 with the addition of FL-N becomes organized and condensed (red puncta background). Image credit: Oregon State University College of Science.
The nucleocapsid protein (or N protein) of the virus is the main target for disease-fighting interventions due to the crucial functions it performs for the infection cycle of the novel coronavirus and since it mutates at a relatively slow pace.
Vaccines and drugs developed based on the work of the N protein have the potential to be highly effective for a longer time—that is, they are less vulnerable to resistance.
Of the SARS-CoV-2 proteins, the N protein is the biggest partner of the viral RNA. The RNA carries the genetic instructions used by the virus to make living cells, like human cells, produce more of itself, and the N protein attaches to the RNA and safeguards it.
The study results were published in the Biophysical Journal and are a crucial jump-off point for more research on the N protein and its interactions with RNA to gain a deeper understanding of the mechanisms of infection, transmission, and control of SARS-CoV-2.
This study was headed by Elisar Barbar, professor of biochemistry and biophysics at Oregon State, and PhD candidate Heather Masson-Forsythe with assistance from undergraduate students Joaquin Rodriguez and Seth Pinckney.
The team used a wide array of biophysical techniques that quantify changes in the shape and size of the N protein when attached to a fragment of genomic RNA—1,000 nucleotides of the 30,000-nucleotide genome.
The genome is rather large for a virus and requires many copies of the N protein to stick to the RNA to give the virus the spherical shape that is necessary for the virus to make more copies of itself. Our study helps us quantify how many copies of N are needed and how close they are to each other when they stick to the RNA.”
Elisar Barbar, Professor of Biochemistry and Biophysics, Oregon State University
According to Barbar, it is rare to use nuclear magnetic resonance to perform biophysical analyses of the N protein with huge segments of RNA. This is due to the challenges in preparing the partially disordered N protein and long RNA segments, both of which are vulnerable to aggregation and degradation.
However, such studies are a specialty of the Barbar lab. In general, studies by other researchers have been restricted to much smaller pieces of RNA and smaller pieces of the N protein.
Instead of just analyzing the RNA-binding regions of the N protein on their own, the 1,000-nucleotide view enabled the researchers to know that the protein attaches much more powerfully when it is a full-length dimer—two copies bound to one another—and to determine the regions of the N protein that are crucial for RNA binding.
The full protein has structured parts but is actually really flexible, so we know that this flexibility is important for RNA binding. We also know that as N proteins start to bind to the longer RNA, the result is a diverse collection of bound protein/RNA complexes as opposed to one way of binding.”
Heather Masson-Forsythe, PhD Candidate, Oregon State University
Thus, drugs that impede the flexibility of the N protein would be one prospective area for pharmaceutical researchers, added Masson-Forsythe. Another likelihood would be drugs that interrupt any of those protein/RNA complexes that are found to be highly crucial.
Source:
Journal reference:
Masson-Forsythe, H., et al. (2021) Multivalent binding of the partially disordered SARS-CoV-2 nucleocapsid phosphoprotein dimer to RNA. Biophysical Journal. doi.org/10.1016/j.bpj.2021.03.023.