To gain a better understanding of how bacterial RNA gives rise to protein and, at the same time, target these processes to develop novel antibiotics, scientists are now focusing on the special way this process takes place in bacteria.
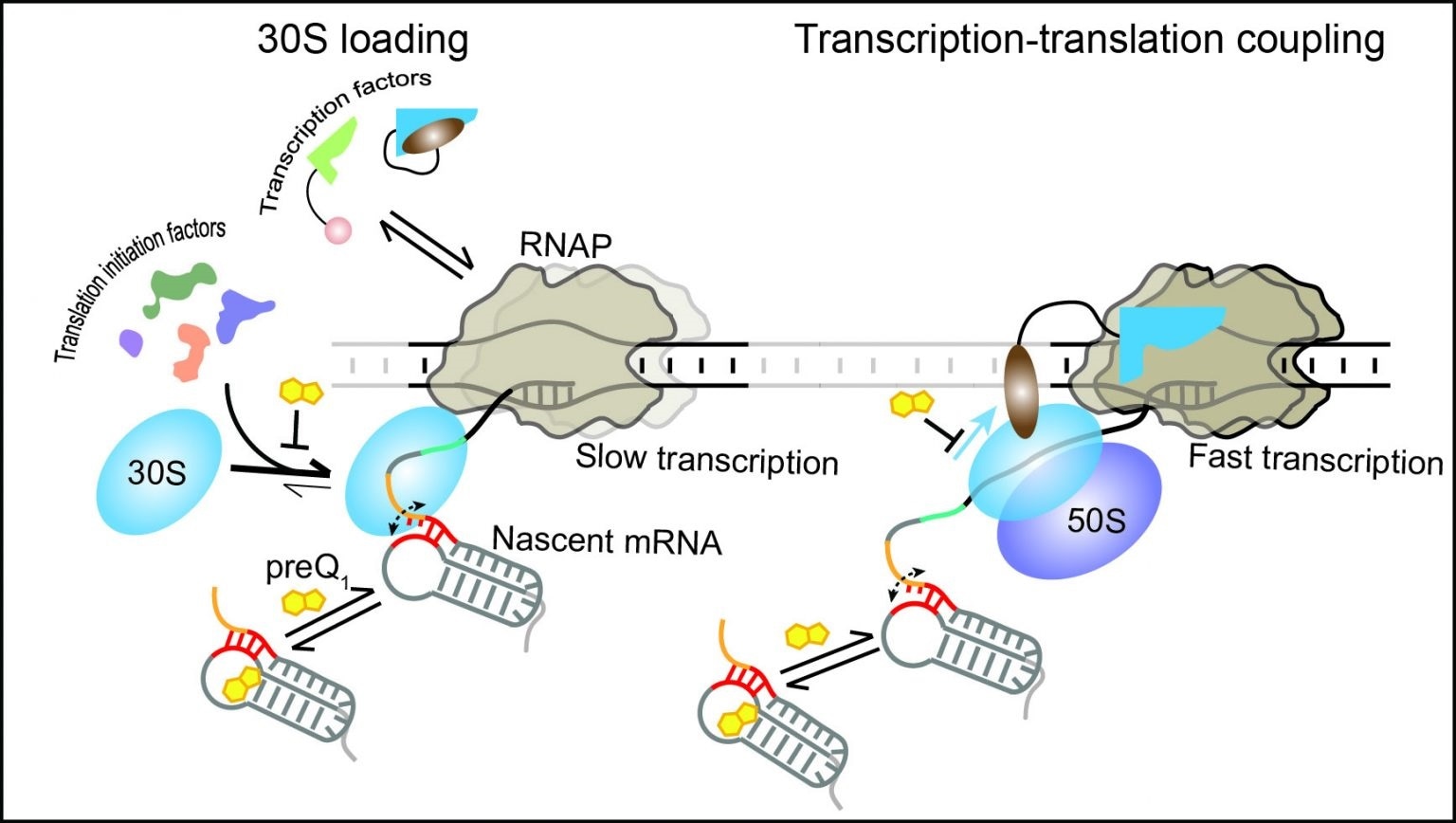
The 30S subunit of the ribosome can dynamically bind to the nascent mRNA as soon as its binding site emerges from the RNAP. Transcription factors and translation initiation factors assist in the initial binding and retention of the 30S subunit on the mRNA, resulting in the stabilization of an early initiation complex. During translation, the ribosome can follow the leading RNAP, establishing transcription-translation coupling and maintaining optimal transcription speed. In the presence of the ligand preQ1, transcription-translation coupling is disrupted, surprisingly leading to slow transcription. Image Credit: University of Michigan.
In the case of eukaryotic cells, transcription (that is, the process through which data in a DNA strand is replicated into the messenger RNA) and translation (the process through which a protein is produced by the ribosome from the mRNA) are two consecutive steps.
In the case of bacteria, both these processes take place at the same time: As the RNA polymerase produces the RNA, the ribosome comes in to form the proteins.
Such synchronicity enables the supposed “transcription-translation coupling,” in which the initial ribosome can instantly track and bind with the transcribing RNA polymerase. This is a new field of study that could provide a better understanding of processes that are unique to bacteria and could be focused on with great specificity during the development of new antibiotics.
Researchers from the University of Michigan (U-M) have now directly visualized the formerly hidden RNA regulatory mechanisms inside these couplings. The outcomes, which were jointly headed by postdoctoral fellows Surajit Chatterjee and Adrien Chauvier from the U-M Department of Chemistry and the U-M Center for RNA Biomedicine, have been published in the Proceedings of the National Academy of Sciences.
These latest findings can have major implications for the upcoming development of new antibiotics that can potentially target the coupling mechanism rather than target the translation or transcription processes independently.
With RNA emerging as a major factor in our daily lives—note the SARS-CoV-2 viral genome and the mRNA vaccines to combat its replication—we are at a crossroads where the interplay between RNAs and proteins in their ubiquitous complexes becomes an attractive prospective target for the medicines of the future, including to fight drug-resistant bacterial strains,”
Nils Walter, Study Senior Author and Professor of Chemistry, University of Michigan
The team specifically noted that modulating the translation of budding mRNA influences the downstream production of the mRNA itself. Upon stopping or delaying the translation, the rate of transcription is slowed down to prevent the excessive production of RNA that would only be decomposed in the cell.
To easily modulate the translation efficiency, the investigators manipulated the traits of a structured RNA, known as a translational riboswitch, integrated close to the beginning of an mRNA of the anthrax bacterium, called Bacillus anthracis. This specific RNA alters its structure when attaching a particular small ligand to decrease the translation process in response to environmental signals.
The latest study demonstrates that the riboswitch—usually assumed to affect only translation—can actually control both transcription and translation processes by manipulating their coupling mechanism.
The researchers used the riboswitch ligand to slow the initiation of the translation process or inhibitors to stop or delay this translation, and they observed that these methods also affect the rate of RNA polymerase.
The researchers then expanded a combination of single-molecule fluorescence microscopy methods to track the dynamic interactions of the translation and transcription machinery during different coupling stages.
The team also devised an exclusive method to directly observe the real-time coupling of transcription and translation and found that the tiny riboswitch regulates the relatively larger translation and transcription machinery. Thus, the new study exceeds and revives the previous structure-based analyses that gave only a glimpse of the already coupled machinery.
According to the investigators, their results lay significant foundations for upcoming RNA studies. The team explained that the question of how other cellular factors play a role in establishing and sustaining the coupling between transcription and translation processes is still vague, raising questions that still need to be explored.
The new study could also provide a better understanding of analogous biological processes in other kinds of pathogenic organisms.
It is fascinating to see how the huge transcription and translation machineries are held by a tiny mRNA for a controlled gene expression process in bacteria.”
Surajit Chatterjee, Postdoctoral Fellow, University of Michigan
Both Chatterjee and Chauvier are senior postdoctoral fellows in the Walter laboratory at the U-M Department of Chemistry. The duo is interested in translational and transcriptional riboswitches, in that order. In the latest analysis, they integrated their interest and knowledge for every aspect of the coupling.
To me, it’s not so much about bacteria, but rather about the biological processes of translation and transcription. Genetic regulation is a timely coordinated process and synchronization is the key for the bacteria to adapt to external threats.”
Adrien Chauvier, Postdoctoral Fellow, University of Michigan
Graduate student Shiba Dandpat and collaborator Professor Irina Artsimovitch from Ohio State University also joined Chatterjee, Chauvier, and Walter in the new study.
Source:
Journal reference:
Chatterjee, S., et al. (2021) A translational riboswitch coordinates nascent transcription–translation coupling. Proceedings of the National Academy of Sciences. doi.org/10.1073/pnas.2023426118.