A team of researchers from the Austrian Academy of Sciences’ Gregor Mendel Institute of Molecular Plant Biology (GMI) has created the first plant embryo gene expression atlas at the single-cell level.
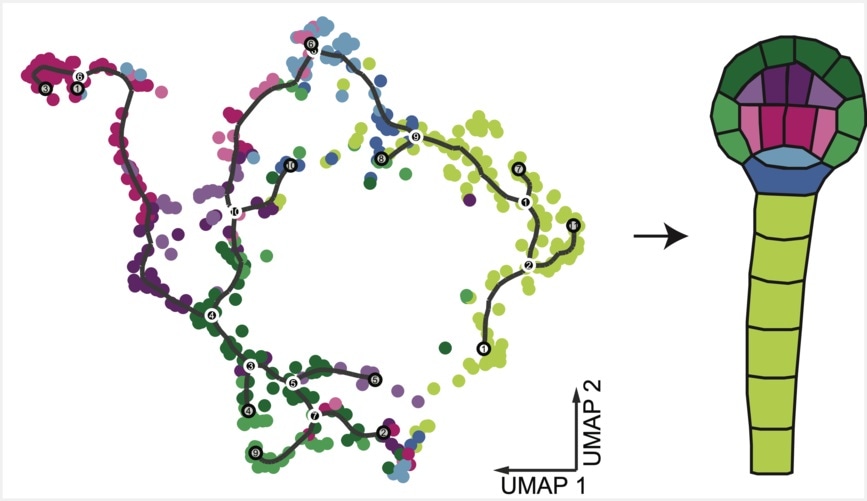
The supervised clustering approach used by Kao et al. resolves nine different cell types in the plant embryo. This hypergeometric test calculates scores for cell types based on marker genes. Each dot represents a nucleus and is assigned to the cell type with the highest calculated score. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. Image Credit: Kao/Nodine/Development/Gregor Mendel Institute of Molecular Plant Biology (GMI)
The study, which was recently published in the Development journal, represents an important step toward understanding the molecular processes that govern how the most fundamental types of plant cells emerge at the beginning of life.
After fertilization, early plant embryos emerge through a fast initial diversification of their component cell types. Consequently, the embryo’s body plan is quickly sculpted by this series of synchronized cell divisions.
Transcriptional activation of the plant genome orchestrates the developmental event in question. But the fundamental cellular differentiation programs have remained hidden for a long time because it was difficult to isolate plant embryos.
As a matter of fact, previous attempts made to create databases of plant embryonic differentiation programs failed to overcome two major challenges: either the datasets were polluted with material from adjacent non-embryonic tissues, or the data obtained lacked cell specificity.
Now, a group of Ph.D. students from GMI Group Leader Michael Nodine’s laboratory has devised a technique for profiling gene expression in Arabidopsis embryos at the single-cell level.
The authors, headed by Ping Kao and Michael Schon from the Nodine group, applied a strategy in which fluorescence-activated nuclei sorting is coupled with single-nucleus RNA sequencing (snRNA-seq). Hence, they were able to sort individual cells in a developing plant embryo and sequence the messenger RNA within individual nuclei. This offers information about the different transcriptomes, or transcription patterns, found in each cell of the plant embryo.
With this method, the researchers were able to overcome the challenges that had stymied earlier attempts to create gene expression atlases in plant embryos. Michael Schon from Nodine’s team instantly identified a striking connection to describe his team’s unique method.
If you put a hamburger in a blender, it still has all the same components in all the same ratios, but you lose critical information about the burger's spatial organization. Elements essential to ‘burger-nesses exist in the organization of the burger's parts, and these elements are lost in a ‘burger smoothie’.”
Michael Schon, Gregor Mendel Institute of Molecular Plant Biology
“Earlier methods consisted of grinding entire plant embryos into a ‘plant embryo smoothie’; these were still useful in telling us the molecular components of the plant embryo and their ratios, but important information about the organism's organization was lost. Our transcriptome atlas is an effort to restore the information that most likely got ‘averaged out' in previous attempts,” added Schon.
Through this method, Nodine’s group was able to demonstrate gene expression patterns that could clearly differentiate the early Arabidopsis embryonic cell types. As a result, the new study paves the way for future research into the molecular basis of pattern formation in plant embryos.
“This is the beginning of an exciting era of developmental biology and approaches like ours promise to help reveal how emerging cell types are defined at the beginning of plant life,” concluded Michael Nodine with a self-assured smile of contentment.
Source:
Journal reference:
Kao, P., et al. (2021) Gene expression variation in arabidopsis embryos at single-nucleus resolution. Development. doi.org/10.1242/dev.199589.