Scientists from KAIST inspected the accumulation of bioplastic granules in living bacterial cells with the help of 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic “polyhydroxyalkanoate” (PHA) employing optical diffraction tomography offers insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
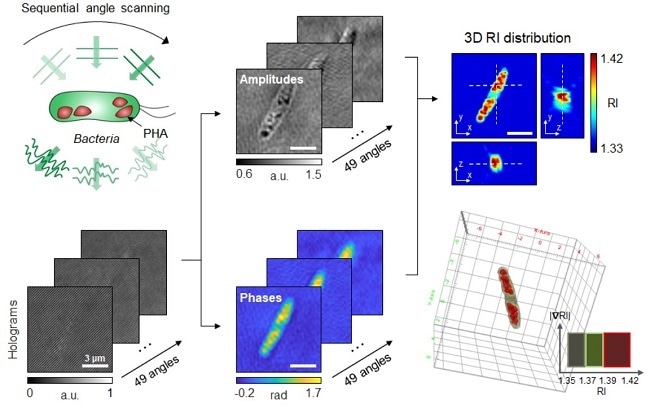
Schematic process of 3D optical diffraction tomography for the bacterial cell accumulating bioplastic polyhydroxyalkanoate (PHA). A cell sample is illuminated at multiple sequential illumination angles (Left, Top). From the raw holograms recorded at individual angles (Left, Bottom), quantitative amplitude and phase information (Middle) is retrieved and the 3D refractive index distribution (Right, Top) is reconstructed. The 3D rendering image of the sample is then obtained (Right, Bottom). Image Credit: Korea Advanced Institute of Science and Technology.
Polyhydroxyalkanoate (PHA), the biodegradable polyester, is being publicized as an environmentally-friendly bioplastic to replace prevailing synthetic plastics. Along with having properties similar to general-purpose plastics like polypropylene and polyethylene, PHA can be utilized in numerous industrial applications like disposable products and container packaging.
PHA is produced by several bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA prevails in the form of insoluble granules in the cytoplasm. Earlier research on identifying in vivo PHA granules were carried out using transmission electron microscopy (TEM), fluorescence microscopy, and electron cryotomography.
The above-mentioned techniques generally depend on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells are to be fixed and sectioned, preventing the investigation of living cells. On the other hand, fluorescence-based techniques call for dye staining or fluorescence labeling.
This being the case, indirect imaging using reporter proteins cannot showcase the native state of PHAs or cells. The invasive exogenous dyes affect the viability and the physiology of the cells. Hence, it was tricky to fully understand the formation of PHA granules in cells owing to the technical limitations. This resulted in numerous mechanism models being proposed based on observation alone.
The group of metabolic engineering scientists headed by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, announced the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography.
The formation and growth of PHA granules in the cells of Cupriavidus necator, the widely analyzed native PHA [particularly, poly(3-hydroxybutyrate), also called PHB] producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were examined comparatively.
The researchers succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level from the reconstructed 3D refractive index distribution of the cells. Specifically, researchers newly introduced the concept of “in vivo PHA granule density.”
By performing the statistical analysis of numerous single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two microorganisms were identified.
In addition, the researchers also recognized the significant protein that plays a vital role in making the difference that allowed the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The scientists also provided 3D time-lapse movies displaying the actual mechanism of PHA granule formation along with cell growth and division. Movies depicting living cells synthesizing and accumulating PHA granules in their native state have never been reported earlier.
This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation.”
Sang Yup Lee, Distinguished Professor, Metabolic Engineering and Synthetic Biology, Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology
“Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future,” adds Professor Lee.
Source:
Journal reference:
Choi, S. Y., et al. (2021) Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individual bacterial cell in its native state. Proceedings of the National Academy of Sciences of the United States of America. doi.org/10.1073/pnas.2103956118.