The genetic material of all eukaryotic organisms is stored in the cell nucleus in the form of DNA. The DNA is first transcribed into messenger RNA in the cell cytoplasm and later translated into protein using ribosomes for it to be used, where ribosomes are small machines capable of decoding messenger RNA to synthesize the relevant proteins.
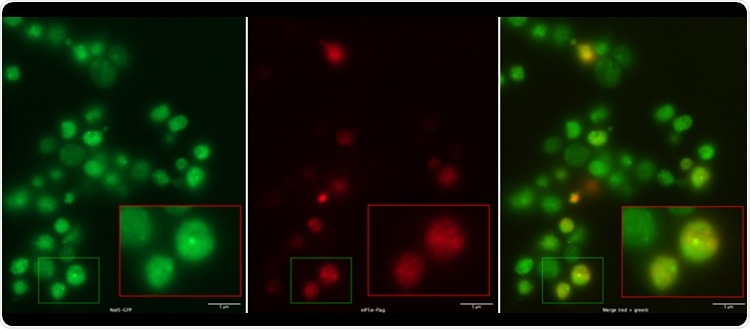
Not5 condensates (on the top, in green) exclude an mRNA translation acceleration factor (visible in red, middle). On the bottom, merged staining: yellow would indicate co-localization, but condensates are green so the mRNA translation acceleration factor is absent. Image Credit: University of Geneva – Laboratoire Collart.
But the speed with which this process occurs is uneven—it must adjust to enable the protein to acquire the correct configuration. Undoubtedly, deregulation of the production rate results in structural defects. The proteins that are incorrectly folded will aggregate, turn unusable, and are mostly toxic for the cell.
By examining the rate of ribosome movement in yeast cells, researchers from the University of Geneva (UNIGE), Switzerland, in association with the University of Hamburg, were able to prove that the rate of protein synthesis is modulated by regulatory factors that change at the rate of translation of messenger RNA into proteins. The findings were published in the Cell Reports journal.
Proteins are 3D structures that need to interlock with one another or interact with partners to act. When a structural defect occurs, the proteins cluster together, turning toxic and possibly pathological. This phenomenon is mostly seen in numerous neurodegenerative diseases, like Alzheimer’s disease or amyotrophic lateral sclerosis.
We already knew that the rate at which proteins are made varies according to need: sometimes fast, sometimes very slow. However, we did not yet know how this mechanism was controlled.”
Martine Collart, Professor, Department of Microbiology and Molecular Medicine, Faculty of Medicine, University of Geneva
Professor Martine Collart headed the study.
Ribosome profiling
To obtain a better picture of the mechanism, the researchers employed a very inventive and yet largely unknown technique: ribosome profiling.
“This methodology makes it possible to determine the position of ribosomes at a given moment in the cell,” states Olesya Panasenko, a researcher in Martine Collart’s lab and head of the “BioCode: RNA to Proteins” Core Facility at the Faculty of Medicine, who is an expert in this technique.
It consists of degrading, at a specific moment, all the RNA that is not protected by the ribosome, to keep only the ribosome-protected fragments (RPFs). We then sequence these RPFs to define how many ribosomes were on the mRNA, and at which positions, at that particular moment. This indicates the speed and efficiency of translation.”
Olesya Panasenko, Department of Microbiology and Molecular Medicine, Faculty of Medicine, University of Geneva
To determine potential differences based on the genetic code, the researchers found the speed and dynamics of protein production in natural yeast cells and genetically modified yeast. At the time of synthesis, small condensates of RNA and proteins show up in the cell, with the task of delaying the rate of ribosome production.
The formation of these condensates depends on the presence or absence of regulatory factors, called Not, which act as decelerators.”
Martine Collart, Professor, Department of Microbiology and Molecular Medicine, Faculty of Medicine, University of Geneva
Their absence leads to acceleration of the process in the wrong places and results in aggregated proteins.
A speed regulated by the genetic code
Accordingly, Not factors link with the ribosome at precise instances in the course of protein synthesis to decelerate the ribosome during translation by condensing the RNA and the nascent protein.
“One may wonder whether this regulatory mechanism is affected during neurodegenerative diseases or with age,” the researchers remark. It is possible that small disturbances, when added together, might eventually have a substantial cumulative effect over time.