According to recent research published out of the Chinese Academy of Sciences and Fresenius Kabi, a multi-enzyme platform was created for sequence-unconstrained traceless protein synthesis and modification with either recombinant proteins or synthetic peptides.
Four biocatalysts work in series to realize the traceless conjugation of synthetic peptides and recombinant proteins, presenting broad application scope. Photo credit: China Science Publishing & Media Ltd. Image Credit: Science China Press.
The observations reveal that enzymes with different functions can be rationally harnessed to provide traceless protein synthesis and functionalization with exceptional flexibility in the choice of both ligation sites and peptide substrates. The research was published in the National Science Review journal.
Protein chemical synthesis and semisynthesis have heavily impacted life science research and pharmaceutical innovation. For around 20 years, the generation of protein synthesis strategies was concentrated on performing the selective ligation of two unprotected peptides in the presence of the plentiful reactive side chains.
The achievable retrosynthetic disconnections of the actual chemical processes are limited to only a few amino acid residues owing to the precise molecular structure of the ligation site for chemoselective capture.
Nature brings forth sophisticated biomolecule systems for catalyzing a tremendous amount of reactions with excellent chemo-, regio- and stereoselectivity. We envisioned that the sequence-independent assembly of native peptides might be feasible by using multiple enzymes that present both strict regioselectivity and broad substrate specificity.”
Dr Bian Wu, Professor, Institute of Microbiology, Chinese Academy of Sciences
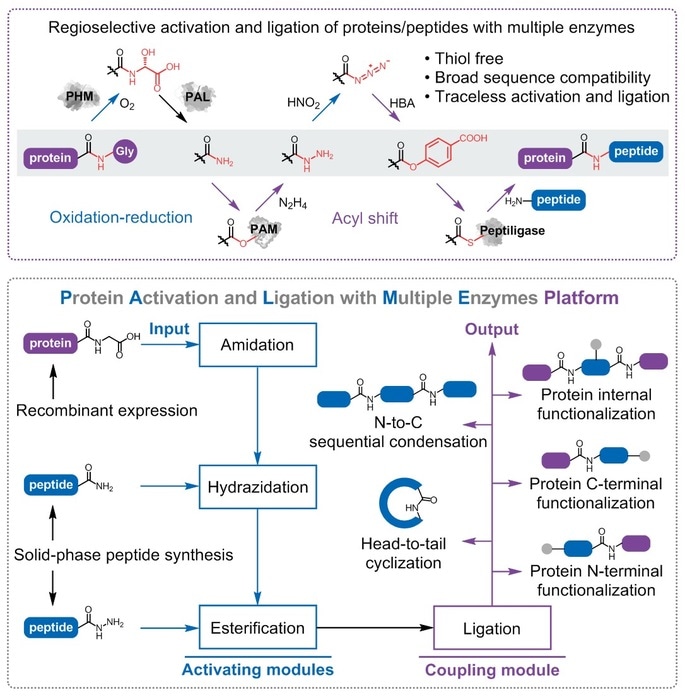
During the mechanism of natural protein synthesis by the ribosome, the association of aminoacyl-tRNA synthetases (activation) and ribosome (ligation) allows accurate assembly of different amino acids without additional protecting groups. Similarly, at least one enzyme for peptide C-terminal activation and an additional enzyme for amide-forming is necessary to enable the conjugation of native peptides without side-chain protection.
For more than 10 years, Wu and colleagues worked on the engineering and mining enzymes connected to peptide processing. Extraordinarily effective and advanced peptide amidase (PAM) for peptide C-terminal functionalization, and the Peptiligase family for peptide conjugation, were created and shown to be appropriate in industrial pharmaceutic peptide manufacture.
In the current research, a bridge combining the two biocatalysts is introduced to construct the whole reaction route. The peptide amide, one among the basic solid-phase peptide synthesis (SPPS) products, is originally altered by PAM to create the corresponding peptide hydrazide. Later, the peptide hydrazide is oxidized and transformed into the peptide ester, which is eventually used as the substrate of Peptiligase for traceless ligation.
This cascade reaction route is also obtainable for recombinant proteins with the help of peptidyl-glycine hydroxylating monooxygenase (PHM) and peptidyl-α-hydroxyglycine amidating lyase (PAL) that can form the des-glycine peptide amide.
The flexibility of each enzyme was analyzed through around 200 model reactions, revealing the magnificent sequence compatibility of the protein activation and ligation with the multiple enzymes (PALME) platform.
Furthermore, to analyze the application scope of the PALME platform, Wu and colleagues produced a group of real-case targets ranging from active pharmaceutical substances to functional modified proteins, including numerous intractable targets—an internally functionalized biomacromolecule and a recombinant protein bearing multiple adjacent Cys residues.
Due to the modular nature, the PALME platform is greatly complementary to other sophisticated methods such as flow chemistry for developing cooperative protein synthesis procedures.
We anticipate that this study will serve as a blueprint for the future development of a widely applicable protocol to access synthetic proteins, and facilitate the development of synthetic biology.”
Dr Bian Wu, Professor, Institute of Microbiology, Chinese Academy of Sciences
Source:
Journal reference:
Li, R., et al. (2021) Traceless enzymatic protein synthesis without ligation sites constraint. National Science Review. doi.org/10.1093/nsr/nwab158.