Researchers from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences devised a simplified and greatly efficient 3'RNA-seq technique, named Simplified Poly(A) Anchored Sequencing (SiPAS), to enhance population transcriptomics in plants.
Comparison of SiPAS with TruSeq. Image Credit: Institute of Genetics and Developmental Biology.
The research was published on September 12th, 2021, in the Plant Biotechnology Journal.
RNA sequencing (RNA-seq) is a major technology for modern biological studies. It shifts numerous genomic studies from a genomic level to a multi-omic level and therefore efficiently increases the understanding of genome biology.
For the past few years, large amounts of genomic data were created in many plant species. The huge amounts of genomic data are generating a vacuum where a great quantity of transcriptomic data requires to be filled to help decode the function of the genome. Greatly efficient RNA-seq technologies are highly demanded in biological research.
The advent of 3'RNA-seq is a great leap of RNA-seq techniques, with active methodological development in the 3'RNA-seq technique. These 3'RNA-seq techniques normally employ custom sequencing format and are not optimized for the standard paired-end 150/250 bp (PE150 or PE250) sequencing. This obstructs their application in large-scale population transcriptomic research.
In the current research, Jing Wang, Jun Xu, Xiaohan Yang, and other researchers in Fei Lu’s research team created an effective gene expression profiling approach named SiPAS. They combined the advantages of reported 3'RNA-seq methods and optimized the use of standard PE150 sequencing format.
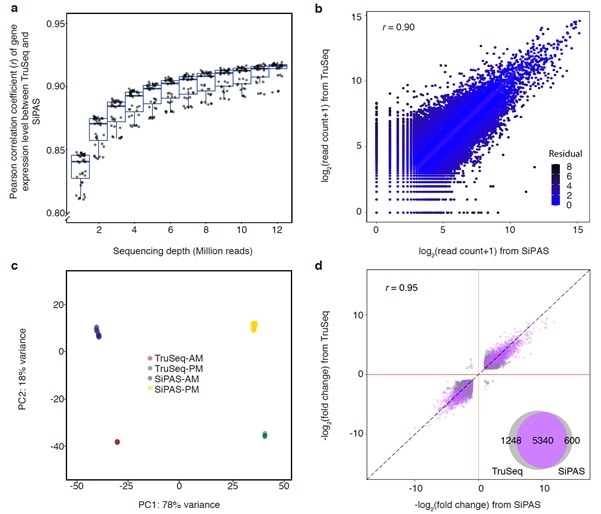
The researchers provided proof demonstrating SiPAS achieves a greater level of accuracy, sensitivity, and reproducibility by analyzing its performance in bread wheat.
The scientists state that the enhanced 3'RNA-seq technique SiPAS offers multiple strengths to promote population transcriptomic research works in plants. The technique is simple and economical with a simplified workflow. Additionally, it is greatly efficient in estimating gene expression and is powerful for RNA degradation.
In general, the strengths of being economical and labor-saving along with a parallel performance with TruSeq enables SiPAS to guarantee effortless use in large-scale population transcriptomic research. It is expected that SiPAS could be employed in numerous species and contribute to a thorough knowledge of plant genomes.
Source:
Journal reference:
Wang, J., et al. (2021) Boosting the power of transcriptomics by developing an efficient gene expression profiling approach. Plant Biotechnology Journal. doi.org/10.1111/pbi.13706.