Please can you introduce yourself and tell us what led you to begin this research?
Bart: I am a physicist who over the years got more and more interested in biology. I am now a Professor of Biophysics at University College London, where I lead a team of researchers who mostly work on microscopy methods and applications to visualize biology at the nanometre scale.
We often use a type of feeling microscopy, called atomic force microscopy (brief: AFM), in which we visualize biological molecules by tracing their contours with a miniature probe, very similar to a blind person reading Braille, just at a much, much smaller scale.
Via collaborators, we had become interested in peptides and proteins that can kill bacteria. This led to microscopy experiments on bacteria, which led to us achieving higher resolution on these bacteria than we had previously thought possible, and which next led to the observation that the surface of E. coli, a very common and well studied Gram-negative bacterium is much patchier than people had previously believed.
Georgina: I am a postdoc in Barts group at University College London, where I also did my Ph.D. My background was in biochemistry but my Ph.D. introduced me to the world of biophysics and the different perspective it provides. I’ve been using AFM to look at biological problems because it yields a very high resolution of live and dividing cell surfaces making it a powerful method for studying dynamic, very small, systems. This gives us a nice way of trying to answer long-standing biological questions that were previously unanswerable.
But, for this research, we started it by accident- while we were looking at other processes on bacterial cells, we saw some features on the surface that none of us could identify. So we decided we’d find out what they were and they were these pores and patches!
What did you find out about the outer layers of Gram-negative bacteria?
We already knew that Gram-negative bacteria have an outer membrane that contains many, many proteins as well as fatty molecules with sugary chains (glycolipids) at the surface. At the nanometre length scale, many biological membranes are fluid. That is, molecules can move rather freely within the membrane.
In contrast, we found that proteins in the E. coli outer membrane formed a rather immobile network that at first sight appeared to span the surface of the entire bacterium. On closer inspection, however, we found that this network was interrupted by smooth patches that are free of common outer membrane proteins. In physics language, we can see that these patches represent domains that are “phase-separated” from the protein network.
So the main conclusion was that at the bacterial surface (at least that of E. coli), there are in fact two different types of surfaces – protein-enriched and glycoprotein-enriched – that matter when we consider, e.g., how bacteria are protected against various antibiotics that target their surface.
Image Credit: fusebulb/Shutterstock.com
Why is it so important to study these outer layers of Gram-negative bacteria?
Several Gram-negative bacteria are among the bacteria that are hardest to kill by antibiotics. The World Health Organisation has identified them as a critical priority for research on and development of new antibiotics. As bacteria develop resistance against currently available antibiotics, we are simply left with very few options to treat even the most common bacterial infections. And Gram-negative bacteria leave us with even fewer options.
To a large extent, this is due to the protective outer membrane that surrounds Gram-negative bacteria. Quite generally, by understanding better how this protective layer is built and organized, we hope to be able to identify vulnerabilities, to inform and guide antibiotic development.
How did you use a needle to visualize the molecular structure of a bacterial surface and obtain these novel images?
As mentioned above, the technology we use is called atomic force microscopy (brief: AFM), which follows the contours of a (here: bacterial) surface in a similar way as a blind person reads Braille, just with a very, very small finger. There is not one key breakthrough that allowed us to take this image; instead, it resulted from the optimization of a whole range of things that went into this, including methods to adhere bacteria to suitable substrates and more technical details on the way we drive this “small finger” over the bacteria.
You may compare this with cooking: An exquisite dish depends on a judicious choice and appropriate balance of several high-quality ingredients, rather than on just having added extra salt or pepper.
Why is the patchy membrane of use to the bacteria and how could these properties be manipulated in the development of antibiotics?
Those are exactly the questions we can start asking now we know that the bacteria are patchy. At the moment we could propose that protein-free patches make it easier to insert new proteins in the membrane as the bacteria grow, since elsewhere the membrane may simply be too densely packed with proteins already. For now, however, that is no more than speculation, and we need many more experiments to validate (or invalidate) that proposition.
As part of this study, we have already identified a type of patches that emerge when the bacteria are missing some key proteins that help to maintain the outer membrane. The appearance of those patches correlated one-to-one with bacterial sensitivity to an antibiotic that does not normally kill Gram-negative bacteria. That suggests that there may be ways by which the patchy nature of the bacteria can represent previously hidden vulnerabilities, yet again, it needs further research to prove or disprove that.
Your findings could help to explain how bacteria can allow for rapid growth whilst maintaining a protective barrier. How has this research improved our understanding of Gram-negative bacteria?
Quite generally, this research has helped us to define the architecture of an essential part of Gram-negative bacteria.
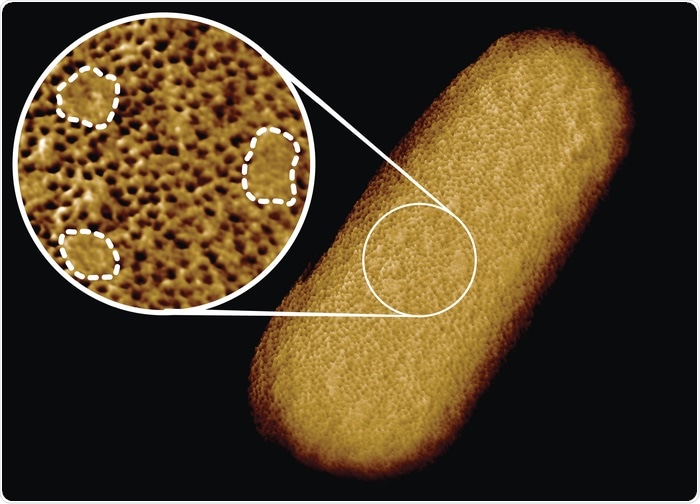
Image Credit: Benn et al. UCL
What is the relevance of antimicrobial resistance in Gram-negative bacteria in today’s society?
Gram-negative bacteria such as A. baumannii, P. aeruginosa, and Enterobacteriaceae (which include Salmonella and E. coli) top the list of bacteria considered the greatest threat in terms of developing (or having developed) antimicrobial resistance, even more so than the well known resistant hospital bug MRSA. This means that infections with resistant strains of these bacteria are increasingly hard to cure, such that infections that were easily treated in the past may be or soon become life-threatening unless we discover new antibiotics.
What other implications could these findings have?
Again quite generally, the outer membrane protects bacteria against harsh environments. Applications of bacteria in e.g. degradation of plastics can also depend on this type of protection. So by understanding the outer membrane better, we may try to better select and/or engineer bacteria as biotechnological tools.
The technology used in this research has created the sharpest images of living bacteria ever recorded. How could this technology be used elsewhere in biosciences to improve knowledge?
It is already extensively used. For example, we have used it in the past to see how DNA can adopt structures that deviate from the canonical Watson-Crick double helix. There is a wide body of research using this technology to study membrane proteins, and it is a common tool to assess the mechanical properties of cells and tissues.
Related to bacteria, colleagues of ours in Sheffield are using similar techniques to study the structure of the bacterial cell wall, which in Gram-negative bacteria lies immediately below the outer membrane, and which represents another target for antibiotics.
Where can readers find more information?
Our own knowledge of those sources relies on the scientific literature, which may not be most easily accessible for a general audience (without us wishing to sound pedantic!).
Quite generally, the Internet provides decent information on general aspects of atomic force microscopy, bacteria, and antimicrobial resistance, which does – at least from what we have seen – is not too much polluted by misinformation.
About Professor Bart Hoogenboom
I am a Professor of Biophysics at University College London. I studied in Groningen (NL) and next did a Ph.D. on superconductivity obtained at the University of Geneva, where I received the 2003 Prix Walthard for combining excellence in sports (middle-distance running) and studies. During subsequent research at the University of Basel, I developed microscopy techniques that facilitated atomic-resolution imaging of surfaces in an aqueous solution.
Since 2007, I have led a lab in London, where we have had a world-premiere by resolving the DNA double helix on a single molecule and, among various other findings, determined how various bacterial and immune proteins get together to punch holes in cellular membranes. For our microscopy work, I received the 2017 Medal for Scanning Probe Microscopy of the Royal Microscopical Society.
About Georgina Benn
I graduated from King’s College London with a BSc in Biochemistry in 2017. I next moved to University College London for Ph.D. research using atomic force microscopy to study the surfaces of live bacteria with nanometre resolution, under the joint supervision of Bart Hoogenboom at University College London and Max Ryadnov at the National Physical Laboratory.
I have recently submitted my Ph.D. thesis and am currently a postdoc.