Viral pathogens such as the SARS-CoV-2 coronavirus alter the interior structure of the cells they infect. These alterations take place at the individual cell components—the organelles—and can offer insight into the development of viral diseases. To visualize them, highly robust imaging techniques are required, however, such techniques are very time- and data-intensive.
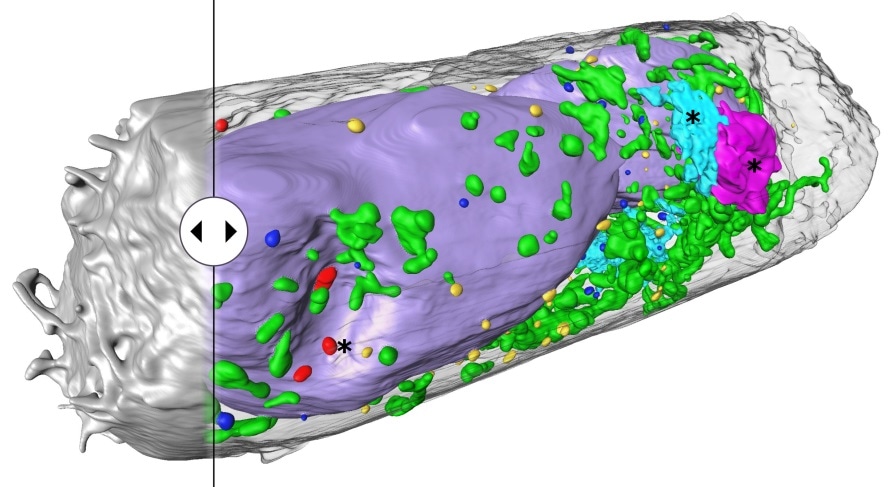
Human lung epithelium cell 24 hours after SARS-CoV-2 viral infection. Hijacked cellular organelles are labelled with an asterisk. Image Credit: © Venera Weinhardt (Centre for Organismal Studies).
A team of German-American scientists guided by Dr. Venera Weinhardt at the Centre for Organismal Studies (COS) of Heidelberg University recently optimized a special X-ray process—called soft X-ray tomography—to provide high-resolution three-dimensional images of entire cells and their molecular structure within a few minutes.
Scanning electron microscopes are preferred in cell imaging because they provide extremely sharp nanoscale images. But this technology takes a good week to scan an individual cell. It also generates an enormous amount of data that is daunting to analyze and interpret. Using soft X-ray tomography, we get usable results within five to ten minutes.”
Venera Weinhardt, Post-Doc, Lawrence Berkeley National Laboratory
According to molecular virologist Professor Dr. Ralf Bartenschlager, whose department at Heidelberg University Hospital associated with Dr Weinhardt on imaging cellular changes linked with viral infections, high throughput is particularly vital for investigating many cells.
Dr. Bartenschlager further states that mostly in tissues only some of the cells get infected and those cells could offer information on the alterations that occur directly from the infection. However, it is not possible to analyze these cells with a scanning electron microscope.
The procedure called soft X-ray tomography (SXT) was used earlier to successfully identify single virus particles—virions—of various kinds of viruses and the changes in the cells linked to them. Recently the scientists employed the technique to analyze cell cultures infected with SARS-CoV-2 from kidney and lung tissue. Soft X-rays enabled them to capture images of complete cells and their structure in three dimensions in around 5–10 minutes.
The scientists could also identify clusters of SARS-CoV-2 particles on the surfaces of the cell and also identify virus-linked alterations in the cell’s interior. Structures that possibly facilitate the replication and spread of the virus were revealed.
Dr Weinhardt says that the group’s success was largely due to the technology enabling them to investigate fixed cells, i.e., cells that were been chemically treated to deactivate the virus. Generally, in soft X-ray tomography, similar to electron tomography, flat lattice structures are employed as holders.
Upon tilting, the thickness of the samples could change. This makes some cell structures blurry. At times the flat shape of the holder obstructs the cells from being scanned at all angles resulting in “Blind” spots. Another problem is that the samples could attach to the lattice or disperse, requiring numerous tomograms to visualize the whole cell.
To get around this problem, we switched over to cylindrical thin-wall glass capillaries to hold the samples. During microscopy, the samples can be rotated a full 360 degrees and scanned from all angles.”
Venera Weinhardt, Post-Doc, Lawrence Berkeley National Laboratory
The scientists are currently absorbed in further refining sample preparation techniques, automating the analysis of the 3D image data, and generating a lab version of a soft X-ray microscope.
Source:
Journal reference:
Loconte, V., et al. (2021) Using soft X-ray tomography for rapid whole-cell quantitative imaging of SARS-CoV-2-infected cells. Cell Reports Methods. doi.org/10.1016/j.crmeth.2021.100117.