Skoltech’s iMolecule group has created an artificial intelligence-driven solution that employs data on the structure of RNA or DNA molecules to detect sites on them where possible drug candidates could interact. Drug companies can find novel treatments—including antiviral agents—far more quickly and efficiently if they are aware of these binding sites.
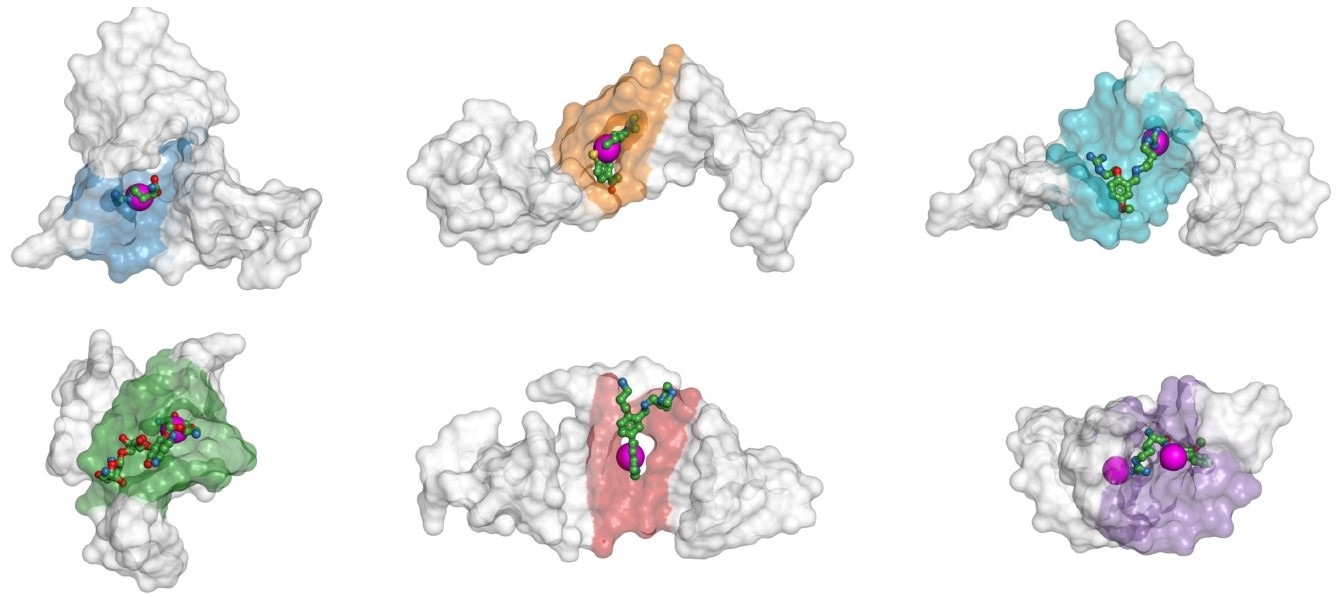
The gray shapes are different spatial configurations of the same RNA sequence of HIV, which is targeted by antiviral medications, such as the compounds shown as stick-and ball skeletons. A neural network presented by Skoltech researchers predicts binding sites as purple spheres, which visibly coincide with the true binding sites highlighted as areas shaded in blue, orange, cyan, etc. Image Credit: Igor Kozlovskii, Petr Popov/Nucleic Acid Research: Genomics and Bioinformatics.
The new method is also more precise than previous methods since it takes into consideration how a nucleic acid molecule’s shape affects which binding sites are visible. The research was published in the journal Nucleic Acid Research: Genomics and Bioinformatics.
Most drugs target proteins and pharmacologists have generally seen RNA as just a mediator between DNA—human genome and—the functional proteins it encodes. While around 85% of the genome is translated into RNAs, only a tiny percentage of those RNAs encode proteins.
Noncoding RNAs, on the other hand, fold into different forms called conformations to activate or inactivate certain genes or perform other functions. While noncoding activities can also have a pathogenic dimension, RNA and possibly DNA sequences are becoming more widely recognized as potential drug targets.
Nucleic acids—DNA and RNA—can participate in signaling, for example, and we could target that or any other process they are involved in. This could be a promising strategy for undruggable protein targets, for example, disordered proteins or proteins that lack convenient binding sites. And then there’s also pathogenic RNA foreign to the body, for example in viruses, such as SARS-CoV-2 or HIV.”
Petr Popov, Assistant Professor, Skolkovo Institute of Science and Technology
Petr Popov is the principal investigator of the study.
Pharmacologists need techniques for screening enormous libraries of chemical compounds to identify which of them interact with nucleic acids and where the specific binding sites are to unlock the possibilities of all those potential therapeutic targets.
Popov explained, “We created this new solution by adapting our prior work with proteins. Nucleic acid three-dimensional structures are encoded as high-dimensional tensors. Once this is done, a computer vision algorithm ‘looks’ at the tensors and highlights the areas in the structure that it thinks could serve as binding sites.”
“After the conformation and the binding site have been detected, a more focused drug discovery campaign can be initiated. So, our work is a small step toward rational drug discovery in contrast to the blind screening, which becomes less reliable with growing chemical libraries,” added Popov.
The shape of RNA and DNA molecules adds a twist to the equation. They have a tend to twist and tangle into various forms. These conformation changes alter the properties of the molecules, including what binding sites are exposed. Conventional methods look only at nucleic acid sequences and neglect conformation, making them inherently unreliable.
“Most earlier methods only worked with RNA, and specifically, with a single chain. Ours works with DNA and with two or more chains. We can even see additional sites that arise when multiple molecules become entangled” said Igor Kozlovskii, a PhD student and the first author of the study.
A great example of what makes working with methods that ignore conformation problematic is the dominant type of HIV. It has an RNA region targeted by many agents. But even though the nucleic acid sequence is the same, when that molecule changes conformation, this is known to have an effect on which agents work or don’t. Our neural network predictions actually reproduce this effect, which means they are reliable.”
Igor Kozlovskii, Study First Author, Skolkovo Institute of Science and Technology
The new solution has an unusual application: it allows you to use the procedure “in reverse.” Instead of detecting binding sites on a prospective target, the program may focus on a problematic agent, a tiny hormone-like molecule that is causing a disorder, and distract it.
So, we want to bind those small molecules with something. To do it, we need to reverse-engineer a short nucleic acid fragment, called aptamer, that would serve as a decoy for the hormone or other molecule of interest. Naturally, an aptamer must contain a binding site, and our solution can be applied to design aptamers with improved binding properties.”
Petr Popov, Assistant Professor, Skolkovo Institute of Science and Technology
Source:
Journal reference:
Kozlovskii, I & Popov, P (2021) Structure-based deep learning for binding site detection in nucleic acid macromolecules. NAR Genomics and Bioinformatics. https://doi.org/10.1093/nargab/lqab111