Reviewed by Danielle Ellis, B.Sc.Feb 14 2022
Chinese scientists explained their use of genome editing to produce significant disease resistance without any growth problems in a study published online in Nature. This result marks a significant theoretical and technological advancement in the use of susceptibility genes in the breeding of disease-resistant plants.
Genome editing-induced chromosomal rearrangement in wheat confers MLO-associated powdery mildew resistance without growth penalties. Image Credit: Shengnan Li
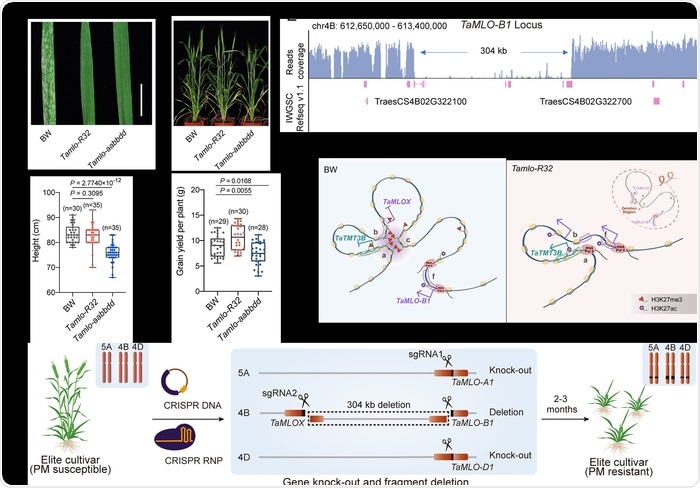
Caixia Gao’s team from the Institute of Genetics and Developmental Biology and Jinlong Qiu’s team from the Chinese Academy of Sciences’ Institute of Microbiology have created Tamlo-R32, a novel wheat mutant with strong resistance to powdery mildew disease and no growth or yield abnormalities.
Plant diseases are responsible for the loss of up to 30% of all crops produced each year, posing a danger to global food security. To establish effective infection events, pathogens often use susceptibility genes (S genes) in plant hosts. Although S gene alterations result in broad-spectrum, long-lasting pathogen resistance, S gene knockdown frequently causes unwanted pleiotropic consequences, restricting its application in disease-resistance breeding.
Wheat (Triticum aestivum) is a key staple crop that feeds almost one-third of the world’s population. Powdery mildew is one of the most common diseases damaging wheat crops across the world.
In 2014, the two research teams generated a wheat variety resistant to powdery mildew disease by genetically manipulating the wheat’s Mildew resistance locus o (MLO), a wheat susceptibility gene. Despite its high disease resistance, this mutant grew poorly when compared to wild-type wheat.
The Tamlo-R32 mutant has a significant 304-kilobase pair deletion in the TaMLO-B1 locus, as well as two premature stop codons in the TaMLO-A1 and TaMLO-D1 loci, according to the researchers. Notably, the substantial loss in the wheat B genome disrupted the local chromatin environment, resulting in ectopic TaTMT3B activation.
The significance of TaTMT3B in reducing the growth and yield penalties associated with MLO disruption was discovered using compensation and knockout studies.
According to the researchers, TMT3 function is preserved in the model plant Arabidopsis thaliana, indicating that it has broader potential across many crop species.
The researchers employed standard breeding procedures to cross the Tamlo-R32 mutant with elite wheat cultivars to effectively incorporate the improved disease resistance features into the elite cultivars, extending the applicability of their study results into agricultural disease resistance breeding.
The researchers also proposed a simple and quick method for leveraging multiplexed CRISPR genome editing technologies to directly construct the matching genetic perturbations in elite wheat types, allowing for the generation of broad-spectrum powdery mildew resistant wheat in about 2–3 months.
This study demonstrates the possibility of stacking genetic modifications to abolish recessive allele-related development abnormalities. Finally, by using S gene-mediated disease resistance, the study offers a novel viewpoint on the application of multiplexed genome editing to breed for disease-resistant and high-yielding crop types.
Source:
Journal reference:
Li, S., et al. (2022) Genome-edited powdery mildew resistance in wheat without growth penalties. Nature. doi.org/10.1038/s41586-022-04395-9.