Researchers have found that marine invertebrates’ genomes are unexpectedly stable over long periods of time. This new study examines the chromosomes of distantly related animal species, such as sponges, jellyfish, scallops, and the invertebrates most closely related to humans, and discovers that they are very similar. The study was published in the journal Science Advances.
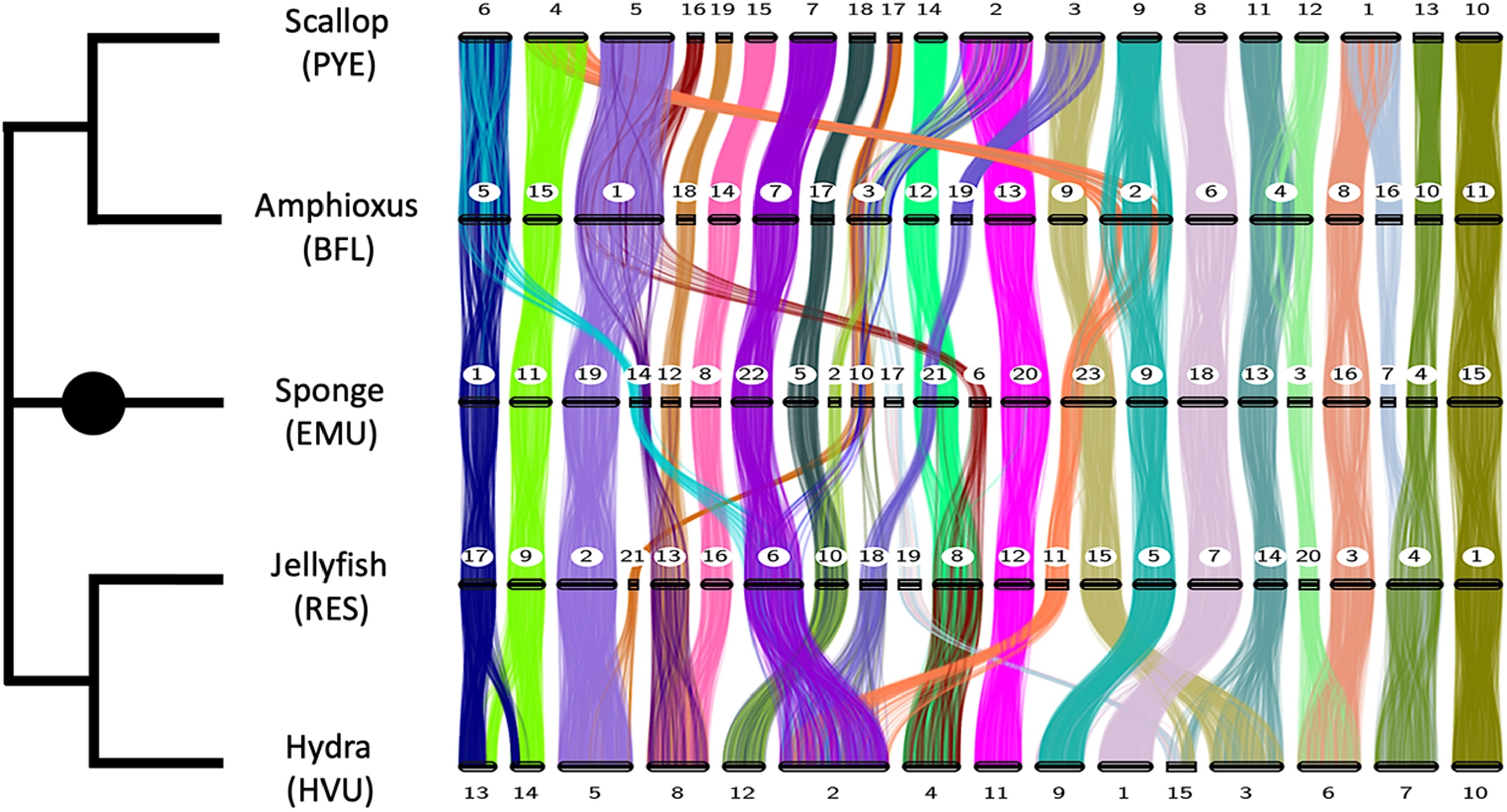
The numbered horizontal bars represent the chromosomes of five species. Each colored strip shows how different sections of gene groups correspond or vary in their location within the different genomes. Two or more colors converging on a chromosome (as can be seen four times with the scallop) indicate that mixing has occurred between two ancestral chromosomes or chromosome sections. Image Credit: The image appeared in the publication in Science Advances.
Consider a genome to be the DNA-coded instruction manual found in each cell. It holds all of an organism’s hereditary information for its functionality. The chromosomes are separated into chapters, which are further split into pages as genes.
Over deep time—and by that, I mean at least 550 million years—due to random mutations, the order of genes within chromosomes become scrambled, kind of like mixing up pages within a chapter of a book. And more dramatically, sometimes we find that two chromosomes have come together and become mixed, as if the chapters were merged and shuffled.”
Daniel Rokhsar, Study Last Author and Professor, Okinawa Institute of Science and Technology Graduate University
Rokhsar is also a principal investigator at the Molecular Genetics Unit at Okinawa Institute of Science and Technology Graduate University (OIST). He further added, “But overall, we found a remarkable amount of stability. Even though the last common ancestor of these three groups lived over half a billion years ago, many of their chromosomes are recognizably similar in the sense that they contain the same groups of genes.”
The genomes of three large groups of animals were evaluated: sponges (simple organisms with no muscles or nerves), cnidarians (particularly jellyfish and hydra), and bilaterians (scallops and amphioxus). These genomes had either been analyzed before or were reported for the first time in this study.
Despite the fact that several of these creatures had previously had “draught” copies of their genomes sequenced, this initial research did not allow for the analysis of general chromosomal arrangement.
Scientists can now put the pieces together and examine how genes are structured into lengthy threads thanks to advances in genetic technology. The hydra’s chromosomes were recreated for the first time, the amphioxus’ chromosomes were significantly improved, and a broad comparative analysis was done in this work.
The international research team, which included scientists from OIST, University of Vienna, University of California campuses in Berkeley, Irvine, and Santa Cruz, Ludwig Maximilian University of Munich, and University College London, discovered strong similarities between the chromosomes of the five animals and affirmed that these resemblances were also existent in other animal genomes.
Researchers discovered patterns of chromosomal fusion that were unique to certain animal sub-groups in several circumstances. The researchers discovered four ancient fusions carried by scallops and many other mollusks, which looked similar to a fusion found in a draught genome of a marine worm.
We see that genes can be on the same chromosome in different species but often in a different order. In rare cases where two chromosomes fuse together and then get mixed up by scrambling across the newly fused chromosome, this fusion can’t be undone, and serves as a permanent marker of the evolutionary history of the chromosome. It’s like shuffling two packs of cards together. We can keep shuffling them, but they will never divide into the two exact packs again.”
Daniel Rokhsar, Study Last Author and Professor, Okinawa Institute of Science and Technology Graduate University
When live creatures share the same fusions, the scientists deduce that the fusions happened in a common ancestor of these species. They’ve now made numerous testable predictions regarding genomes that haven’t yet been identified. For example, the researchers believe that all mollusks and related “spiralian” creatures’ genomes must contain the same set of fusions found in scallops.
These findings reveal a fascinating contradiction.
Mammals have only been around for about 100 million years. But when we compare the genome of two mammals, say, a human and a mouse, the chromosomes look like they have been broken up into a few hundred pieces and then mixed together. The chromosome-scale conservation characteristic that we found in invertebrates is simply not seen in mammals.”
Daniel Rokhsar, Study Last Author and Professor, Okinawa Institute of Science and Technology Graduate University
He theorized that mammalian chromosomes evolved separately than those of many marine invertebrates as mammals have always resided in smaller units. Small groups let random mutations survive, which may explain why chromosomal rearrangements propagate more easily in mammals.
The researchers discovered 29 ancestral chromosomal fragments in total. Moreover, the researchers discovered that some of these segments existed 800-900 million years ago, before the emergence of mammals when all organisms were unicellular or multi-cellular in nature. As a result, some genes have been commuting together for nearly a billion years, but the outcome of these ancient gene couplings is yet unknown.
Source:
Journal reference:
Simakov, O., et al. (2022) Deeply conserved synteny and the evolution of metazoan chromosomes. Science Advances. doi.org/10.1126/sciadv.abi5884.