The science
Microbial communities in nature are made up of a variety of bacterium species. This makes it challenging to isolate and culture individual bacterium species in the laboratory. New tools have been created by researchers to assist. Researchers can use these technologies to modify the genetics of different bacterial species in their communities.
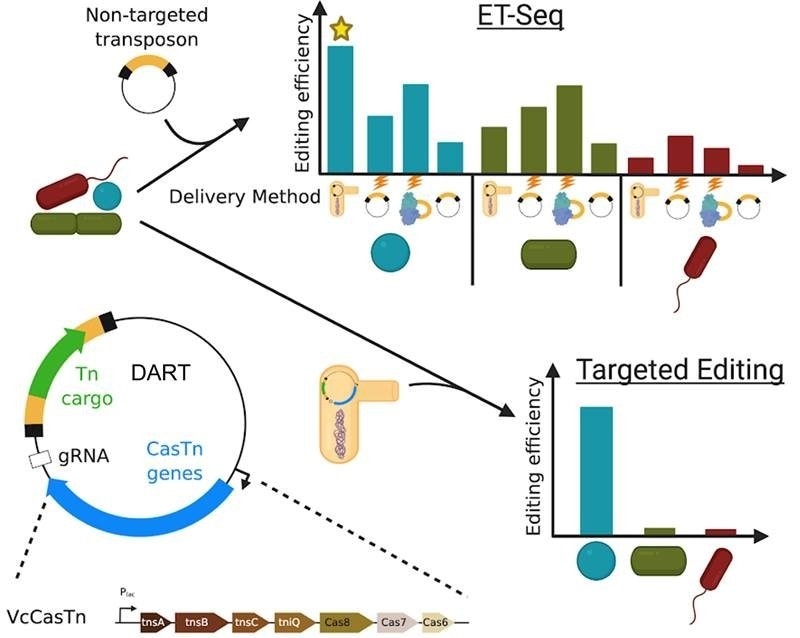
New tools provide targeted genome editing in microbial communities. ET-Seq allows the isolation independent assessment of genetic accessibility of a microbe in community context. The DART vector system allows highly specific targeted DNA insertion. Image Credit: Ben Rubin
One of the technologies, ET-Seq, allows researchers to find bacteria that can be genetically changed precisely within mixed communities. The DART technology, on the other hand, allows scientists to examine the gene function from different species within their communities.
The impact
The majority of bacteria live in complex communities. Scientists must separate individual species to examine them with most modern techniques. Environmental microorganisms, such as soil bacteria, are more difficult to extract and culture in the lab. In addition, the behavior of microbial communities is determined by the contributions of all of its members. This is in stark contrast to the behavior of isolated animals.
Scientists may examine microbial communities without isolating distinct members thanks to the breakthroughs of the ET-Seq and DART technologies. Researchers can trace genetic alterations as the community expands and evaluate gene function in microorganisms that cannot be produced in the lab using both approaches.
Summary
Cultivation and genetic analysis have been the most common methods for studying gene function and microbe behavior. These traditional methods, on the other hand, necessitate isolating and growing bacteria in the laboratory.
Researchers’ knowledge of bacteria that cannot yet be cultured in the lab is severely limited as a result. Furthermore, isolated organisms cannot study the interactions that occur between bacteria when they develop together in a community.
To address these issues, this study created two technologies that may be used to assess functions and connections within laboratory microbiomes. A mobile genetic element (transposon) is delivered into a microbial population via environmental transformation sequencing (ET-seq).
Some bacterial species’ genes are randomly inserted by the transposon. Scientists can detect community members who are converted by the transposon and how frequently they are transformed by sequencing the genomes of all the bacteria in the community. They are able to determine genetically tractable species in this way.
Utilizing DNA-editing all-in-one RNA-guided CRISPR–Cas transposase, those species can be targeted specifically for gene alteration (DART). In a lab environment, the researchers used both strategies to show the enrichment of selected bacterial species, give novel metabolic features, and quantify gene fitness of bacteria in a community context.
These new features will also provide crucial fresh perspectives into the activities of uncultivated microorganisms and the functions of critical genes, metabolites, and proteins, such as in soil carbon cycling and regulating beneficial microbial connections with plants for long-term bioenergy production.
Source:
Journal reference:
Rubin, B. E., et al. (2021) Species- and site-specific genome editing in complex bacterial communities. Nature Microbiology. doi.org/10.1038/s41564-021-01014-7.