In this interview, we speak to Dr. Sam Schaffter about his latest research that looked at creating biological computers using RNA and their role within research.
Please can you introduce yourself and tell us what inspired your latest research into biological computers?
My background is in DNA nanotechnology and computing, two fields that have developed some of the most sophisticated molecular computers to date, enabling mixtures of molecules to compute the square root of numbers and recognize patterns of nearly 100 inputs. Much of my previous work focused on using these powerful molecular computers to control the self-assembly of nanostructures. But what if we could use them to control the behavior of living cells? That question led me to think about the challenges that would need to be overcome to achieve such a goal and addressing some of these key challenges ultimately became the focus of this recent manuscript.

Image Credit: zffoto/Shutterstock.com
So far, DNA-based technology that has been developed in this field has had a variety of drawbacks. Can you tell us about some of these current limitations?
What is considered a limitation is highly dependent on the application. For example, typical DNA-based circuits must be assembled by individually preparing and then mixing the components, and each mixture can only execute a computation one time. For test tube diagnostics this is sufficient, but our goal was to develop a system that could continuously operate in a closed environment, e.g. a cell. The primary challenge of accomplishing this with a typical DNA-based computer is the ability to continuously produce all the circuit components together in the same mixture in which they execute the computations. This is the challenge we sought to overcome with this work.
In your latest research, you have developed a long-living biological computer. Can you describe how you designed this computer and how it works?
We have not yet demonstrated long-lived biological computers in cells. What we have done is design biological computers that can be continuously produced via a biological process called transcription, in which RNA copies of a DNA sequence are synthesized. Continuous production of RNA circuit components via transcription should enable these computers to be long-lived and execute multiple computations in biological systems, including cells. Our current work lays the foundation for this goal.
Our circuits operate using toehold-mediated strand displacement (TMSD), originally developed in DNA nanotechnology. In TMSD circuits, short pieces of double-stranded RNA serve as locked gates that can only release an output signal when the correct input RNA is around. The process of unlocking an output is facilitated by a toehold domain on the gates that recruits the input RNA. Upon binding, the input strand replaces one strand on the gate and releases the output strand into solution. Once released, the output strand can trigger reactions with other gates to execute preprogrammed cascades or computations. Our primary innovation was designing the circuit components so they could be continuously produced together without significant cross-reaction – this is what could allow these circuits to be encoded into a cell’s genome to be continuously operated inside living cells.
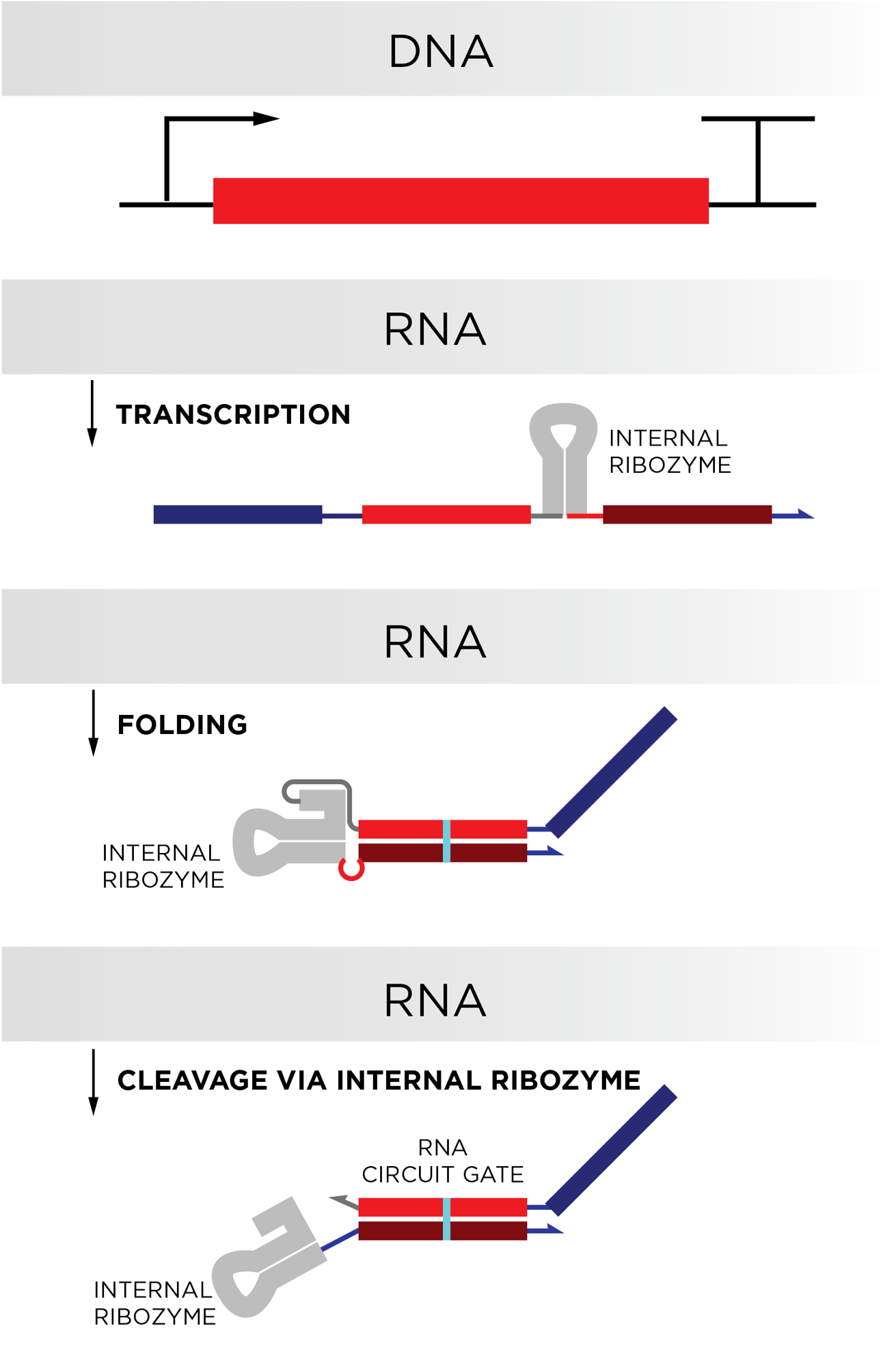
Image Credit: N. Hanacek/NIST
How is the biological computer able to produce useful information? What would this information look like?
These biological computers are particularly useful for processing chemical information. Considering diagnostic applications, a set of specific mRNA sequences could represent biomarkers for disease. The circuits we have developed could be programmed to detect these different RNA sequences and then compute whether the set of sequences corresponds to a specific disease.
Your new biological computer uses RNA as opposed to DNA. Why did you choose to use RNA and what are some of its advantages compared to DNA?
If you were to take the typical DNA-based strand displacement circuit as it is currently prepared and operated in a test tube now and changed the DNA to RNA, there would not be much of a practical difference. The main motivation for using RNA in this work is that it can easily be produced via transcription. So, if we could develop a system in which strand displacement circuit components are encoded as RNAs that can be produced via transcription, then, in principle, the DNA sequence that encodes for the production of the RNA components could be integrated into a cell’s genome and the cell’s natural machinery could continuously produce the circuits. This is the principle of most other biological computers that are used for cellular engineering. But the design rules for how to do this for state-of-the-art strand displacement circuits (typically built with short DNA strands) had not been developed.
How could the biological computer you have made help to revolutionize the way we diagnose and treat disease?
There has been a lot of amazing work developing nucleic acid biosensors and diagnostics that operate in test tubes using both DNA computers and RNA computers, as well as hybrid approaches. The technology we developed has the potential to enable the same capabilities for long-term monitoring and diagnosis in real-time inside living cells. I expand more on this idea in my answer to “With the continued incredible advancements in technology within science, what are you particularly most looking forward to within the field of medical diagnostics?”.
Are there any challenges that you had to overcome during your research? If so, what were they?
To me, the biggest challenge was settling on the transcriptionally encoded RNA gate design, especially how to design the sequences such that the gates robustly fold into the desired structures critical for function. There are many different design choices, including which self-cleaving RNA sequence to use, where to situate that sequence within the gate, and in what order to produce the gate sequences via transcription to reduce misfolding traps during gate production. So, a lot of the initial work was thinking critically about these different design choices and scouring the literature to inform our decisions. The final design we selected seems to work very well, as evidenced by our ability to predictively build circuits with behavior that closely matches our idealized simulations.

Image Credit: CROCOTHERY/Shutterstock.com
With the continued incredible advancements in technology within science, what are you particularly most looking forward to within the field of medical diagnostics?
Early disease detection and prevention with real-time diagnosis of complex disease patterns in living cells. Speculating well into the future, one thing that motivated us early in this work was the idea of engineering biological computers that can continuously monitor for specific disease patterns inside cells. Such computers could serve as “molecular watchdogs” that detect the onset of disease as soon as it arises inside a single cell and potentially trigger corrective action – this may allow us to diagnose and treat disease well before it has a chance to spread to a level that we could detect with current methods.
What are the next steps for you and your research?
One thing we have been working on recently is pushing the boundaries of the RNA gate design to identify failure modes and design heuristics that will give us the best chance for success as we move forward. Specifically, as we work to move this technology into different environments, such as cell lysates and living cells, the optimal gate design parameters could change.
Moving into these new environments poses many new challenges – perhaps most importantly how to reliably measure all the important factors that dictate the operation of the biological computer, such as RNA stability, RNA abundance, RNA structure, RNA:RNA interactions, etc. This research is happening at the National Institute of Standards and Technology (NIST) because these critical measurement challenges need to be overcome for this technology to transition from the laboratory to practical applications that impact people’s everyday lives.
Where can readers find more information?
NIST Cellular Engineering Group website: https://www.nist.gov/mml/bbd/cellular-engineering-group
NIST project page: https://www.nist.gov/programs-projects/cellular-measurement-and-computation�rna-circuits
Dr. Sam Schaffter NIST webpage: https://www.nist.gov/people/samuel-schaffter
Dr. Elizabeth Strychalski NIST webpage: https://www.nist.gov/people/elizabeth-strychalski
Podcast interview with Sam about this and related work: molpigs Podcast | Sam Schaffter
RNA Circuits Paper: Samuel W. Schaffter, Elizabeth A. Strychalski. Cotranscriptionally encoded RNA strand displacement circuits. Science Advances, 2022; 8 (12) DOI: 10.1126/sciadv.abl4354
About Dr. Sam Schaffter
Sam conducted his PhD research in the field of DNA nanotechnology and DNA computing, working in Rebecca Schulman’s group at Johns Hopkins University. He developed synthetic transcription-based networks with dynamics programmed via Watson-Crick base pairing rules. These networks, which operate in test tubes, emulated key functionalities of cellular genetic regulatory networks and thus could serve as a programmable “synthetic genome” for controlling nucleic acid materials and devices, such as DNA nanostructures and DNA-responsive hydrogels. The goal of his research was to engineer synthetic materials capable of sophisticated behaviors seen in biology including hierarchical differentiation or self-healing.
Based on this work, in 2021, he was awarded the Robert Dirks Prize in Molecular Programming (https://dirksprize.caltech.edu/). At the National Institute of Standards and Technology (NIST), under a National Research Council Postdoctoral Research Associateship, Sam is interested in moving nucleic acid computing circuits from the test tube to living cells. Current state-of-the-art DNA-based circuits are only single use and suffer from degradation inside cells, limiting their practical applications. To overcome these limitations, Sam’s current research focuses on transcriptionally encoding RNA-based circuits, equivalent to those developed in DNA computing, that can operate continuously inside living cells. These circuits could be programmed to recognize complex differential gene expression patterns in real-time inside living cells, potentially enabling a new class of living measurement systems.