Reviewed by Danielle Ellis, B.Sc.Apr 22 2022
Tradeoffs between diverse agronomic traits constrain crop breeding to a great extent. Because gene pleiotropy causes many of these tradeoffs, lowering gene pleiotropy may make it simpler to overcome tradeoffs in plant breeding. Few successful techniques, however, have yet to be identified.
 The mechanism of boosting rice yield by overcoming a trait trade-off between tiller number and panicle size. Image Credit: Institute of Genetics and Developmental Biology.
The mechanism of boosting rice yield by overcoming a trait trade-off between tiller number and panicle size. Image Credit: Institute of Genetics and Developmental Biology.
Scientists from Jiayang Li’s team at the Chinese Academy of Sciences’ Institute of Genetics and Developmental Biology employed tiling deletion to alter the cis-regulatory regions of the pleiotropic rice gene Ideal Plant Architecture 1 (IPA1) in a work recently published in Nature Biotechnology. Researchers were able to overcome the tradeoff in rice yield component features in this way, resulting in an effective technique for overcoming the rice production bottleneck.
IPA1, discovered by Li’s team, is a key regulator of rice plant architecture. It produces a plant-specific transcription factor that controls a variety of characteristics of rice growth and development, as well as disease resistance and environmental adaptation.
Gain-of-function alleles in the IPA1 gene result in fewer unproductive tillers, larger grains per panicle, stronger culms, and more robust roots, and hence a higher grain production. As a result, they have become quite popular in rice breeding. IPA1, on the other hand, is a pleiotropic gene that improves grains per panicle while decreasing tillers.

Image Credit: sakhorn/Shutterstock
The researchers altered the cis-regulatory region of IPA1 to alter its expression level in immature panicles, stem bases, and other tissues in order to selectively regulate distinct features, in an attempt to overcome the tradeoff between panicle size and tiller. However, the function of the cis-regulatory region of plants remained mostly unclear due to the limits of prior technical methods.
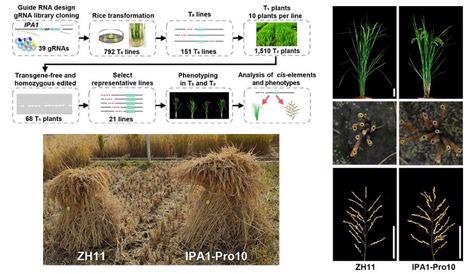
Researchers next used CRISPR/Cas9 to screen for the appropriate cis-regulatory area of cis using tiling deletions. The researchers discovered IPA1-Pro10 in the genome editing library, a line with a 54-bp cis-regulatory region deletion that can improve tiller number and grain number per panicle at the same time.
In paddy fields, IPA1-Pro10 demonstrated larger panicles, higher tillers and plant height, thicker stems and roots, and enhanced yield by 15.9%.
Furthermore, the researchers looked into the molecular mechanism by which the IPA1 cis-element regulates panicle traits, and discovered that An-1, an important transcription factor for domestication, can attach to the GCGCGTGT motif in the 54-bp cis-regulatory area and particularly induced the expression of IPA1 in young panicles, which in turn regulates panicle traits.
This research suggests a viable technique for resolving agronomic trait tradeoffs. It also offers fresh genetic resources for breaking through rice yield barriers. As a result, this research represents a significant advancement in this sector.
Systematic elucidation of essential gene cis-regulatory regions will reveal novel molecular mechanisms and genetic resources for addressing crop breeding constraints in the future.
Source:
Journal reference:
Song, X., et al. (2022) Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nature Biotechnology. doi.org/10.1038/s41587-022-01281-7.